Introduction
Pseudomonas aeruginosa well known opportunistic pathogen, and causes life threatening nosocomial infections due to its intrinsic resistance to many antibiotics and development of increased resistance in healthcare settings. The increasing resistance of potentially pathogenic bacteria to multiple conventional antibiotics is an urgent problem in global public health.1 Among various mechanisms of resistance, ß-lactamase production is the most important, resistance mechanism among P. aeruginosa. Under these ß-lactamases, ESBL producing P. aeruginosa continues to pose a challenge to infection control worldwide. Extended-spectrum ß-lactamases (ESBLs) belong mostly to class A of the Ambler classification scheme and group 2be according to the Bush-Jacoby-Medeiros functional scheme.2, 3
The 2 be designation consists of ‘2b’ denoting that the enzyme is derived from a 2b enzyme (e.g., SHV-1, TEM-1, and TEM-2) and ‘e’ representing the extended spectrum of activity, ESBL producing gram negatives are typically resistant to Penicillin, first and second generation cephalosporins and also third generation oxyimino cephalosporins and monobactams. ESBLs in P. aeruginosa includes the, SHV, TEM, PER, VEB and IBS/GES types4 Early and accurate detection of ESBL producing P. aeruginosa is important for optimal treatment of critically ill and hospitalized patients and also, to control the spread of resistance. Hence, the present study was conducted with an objective to find the prevalence of ESBL producing P. aeruginosa and also to serve as a guide for doctors managing patients by implementing proper infection control measures as well as formulating an effective antibiotic strategy.
Materials and Methods
The present study was conducted in the Department of Microbiology at Muzaffarnagar Medical College, Muzaffarnagar, over a period from January 2014 to July 2015. All 322 isolates of Pseudomonas aeruginosa obtained from various clinical samples received in microbiology laboratory from IPD & OPD were included in the study. The isolates were identified as per the standard microbiological procedures.5 Antimicrobial sensitivity testing was performed on Mueller-Hinton agar plates with commercially available disks (Himedia) by Kirby Bauer disk diffusion method and interpreted as per CLSI guidelines.6
The results of susceptibility test were divided into susceptible and resistant. The isolates with intermediate susceptibility were included in resistant category.
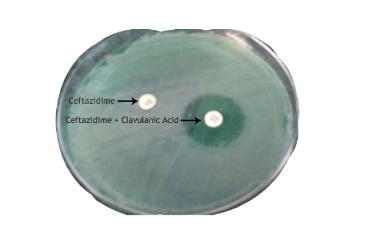
Isolates were considered a potential ESBL producer if the zone of inhibition for ceftazidime was observed to be <22mm. Potential ESBL producer was then subjected for ESBL Phenotypic confirmatory test –Disc Diffusion method as recommended by Clinical Laboratory Standards Institute, 2015.7
Results
A total of 322 non duplicate isolates of Pseudomonas aeruginosa identified during the study period. Out of 322 isolates, 226 (70.19%) isolates of Pseudomonas aeruginosa showed zone of inhibition ≤ 22 mm for third generation cephalosporin, Ceftazidime. of these 26.09% (n = 84) of Pseudomonas aeruginosa isolates were found to be ESBL producers[Figure 1].
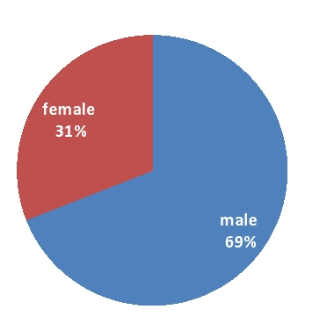
Prevalence of ESBL producing Pseudomonas aeruginosa (84/322) in this study was 26.09%. Out of which 58(69.05%) samples were of male patients and 26(30.95%) were from female patients[Figure 2].
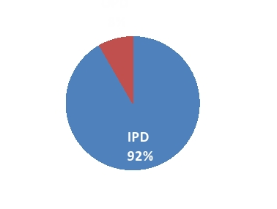
In Patient Department (IPD) 77 (91.67%) and only 07 (8.33%) samples were of OPD patients[Figure 3].
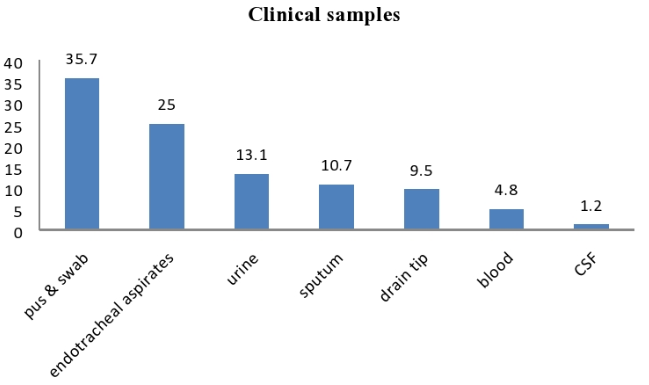
We isolated ESBL positive P. aeruginosa from different type of samples, out of which maximum number were of pus and swabs 30 (35.7%) followed by Endotracheal aspirates 21 (25%), urine 11(13.10%), sputum 09 (10.71%), drain tip 08 (9.52%), blood 04 (4.76%) & cerebrospinal fluid 01 (1.19%)[Figure 4].
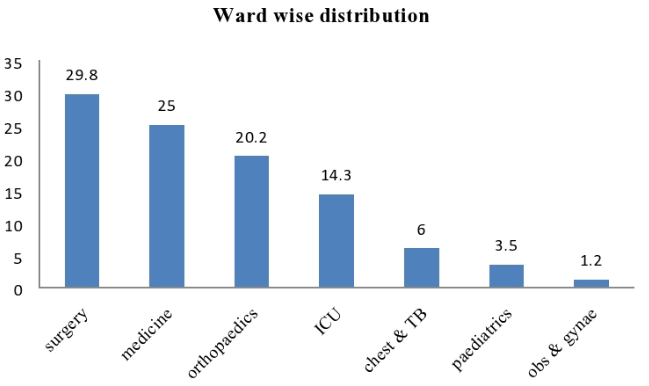
Out of 84 ESBL positive samples of pseudomonas aeruginosa maximum sample received from Surgery 25 (29.76%) followed by Medicine 21 (25%), Orthopaedics 17 (20.24%), ICU 12 (14.29%), Chest & TB 05 (5.95%), Paediatrics 03(3.5%) and Obs & Gynae 01 (1.2%), respectively[Figure 5].
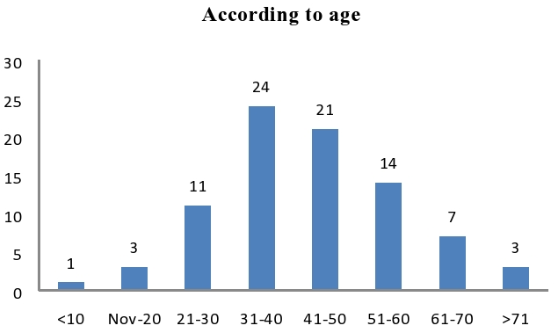
Maximum samples received from middle age group (30-50)[Figure 6].
All ESBL positive Pseudomonas aeruginosa isolates showed high resistance to ciprofloxacin 79(94.05%), Gentamicin 61(72.62%) and tobramycin 60 (71.43%). Resistance was low to combination drugs like cefoparazone +salbactum 17(20.24%) and piperacillin + Tazobactum 14 (16.67%). These strains also showed resistance to carbapenems like Imipenem 15 (17.86%), which were found to be the precious weapon against Pseudomonas aeruginosa infections and this, is an alarming sign. All the isolates showed 100% sensitive to Polymyxin B [Table 1]. All isolates from urine samples showed (100%) resistance to Norfloxacin.
Table 1
Drug resistance pattern of pseudomonas aeruginosa (n=84)
Discussion
Our study showed 84 (26.08%) isolates were ESBL producer which is very similar to other studies, by Prashant et al.,8 and Agarwal et al.,9 which were 22.22% & 20.27% respectively Whereas, high percentage of isolates were ESBL producer (45.19%) by Senthamarai S. et al.,10 42.30% ESBL producer were observed in the study of Varun Goel et al.,11
In the present study, In Patient Department (IPD) 77(91.67%) and only 07(8.33%) samples were of OPD patients. A similar observation was made by Shampa Anupurba et al.,12 and Prashant et al.8 They expressed their view that the duration of the hospital stay was directly proportional to a higher prevalence of the infection, since the rate of isolation of the organisms was higher in indoor patients than in outdoor patients.
Isolates found to be more distributed among age groups between 31 to 50 years of age 41 (27.63%). Similar high prevalence in middle age group is reported by Senthamarai S et al.,10
Pus (35.71%) was the main source of P. aeruginosa followed by Endotraceal aspirate (25%), urine (13.10%), sputum (10.71%), drain tip (9.52%),blood (4.76%) & cerebrospinal fluid (1.19%). Similar results had been reported by Manzoor et al.,13 and Senthamarai S. et al.,10 Although Amandeep Kaur et al.,14 reported more prevalence in urine samples than respiratory samples.
ESBL-producing bacteria are frequently resistant to many other classes of antibiotics, including aminoglycosides and fluoroquinolones. This is due to the coexistence of genes encoding drug resistance to other antibiotics on the plasmids which encode ESBL.15 This fact has also been observed in our study.
Resistance was low to combination drugs like cefoparazone +salbactum (20.24%) and piperacillin + Tazobactum (16.67%) as compare to Suprakash Das et al.,16 and Senthamarai S.10
In our study susceptibility to Imipenem is in accordance with Manzoor et al.,14 however, it is lower compared to the 100% susceptibility found in ESBL-producing gram-negative isolates By Ullah et al.,17 decreased susceptibility to Imipenem is a matter of great concern and indicates the urgent need for improved infection control strategies.
All the isolates showed 100% sensitive to Polymyxin B similar to study done by Kalaivani R et al.,18
Conclusion
Phenotypic combined Disc Diffusion test is widely used due to its simplicity and ease to perform and interpret this test. The present study was done to know the prevalence of ESBL mediated resistance in P. aeruginosa that causes a therapeutic challenge in health care. Most of these isolates were from hospitalized patients which indicate more chances of their nosocomial predominance. The presence of ESBL positivity in out patients is alarming sign for community spread and increase in community-acquired ESBLs.
To reduce the problem of emerging ESBL positive P. aeruginosa, we need team effort from the microbiologists, clinicians and infection control team.