- Visibility 98 Views
- Downloads 19 Downloads
- DOI 10.18231/j.ijmr.2021.007
-
CrossMark
- Citation
Clinical, phenotypic and genotypic profile of carbapenem resistant gram negative infections in intensive care units
Introduction
Carbapenems are beta-lactam antibiotics with a broad spectrum and high efficacy, they are often used as “last-line agents” or “antibiotics of last resort” when patients with infections become gravely ill or are suspected of harboring resistant bacteria and used as drug of choice for infections caused by Gram-negative bacteria that produce extended spectrum beta-lactamases (ESBL).[1] The intensive care unit (ICU) often is called the epicenter of infections, due to its extremely vulnerable population and there is an increase in the use of carbapenems not only for documented infections but also for empirical treatment of acquired infections.[2] Thus, there is a selective pressure for carbapenem resistance in intensive care units of tertiary care hospitals.
Antimicrobial resistance against carbapenems in Gram-negative bacteria can be caused by high level expression of ESBLs accompanied by changed porin expression or by specific enzymes named carbapenemases. Although known as “carbapenemases,” many of these enzymes recognize almost all hydrolyzable β-lactams and most are resilient against inhibition by all commercially available β-lactamase inhibitors.
The present study was done for screening the patients of Intensive care units of a tertiary care hospital for carbapenem resistant gram negative infections, phenotypic detection of the carbapenemases by Modified Hodge test, combined disc tests and genotypic confirmation of resistant gene by PCR as most of the genes conferring resistance are plasmid mediated and bacteria may be harboring more than one such resistant genes, finding out any attributable risk foctors for carbapenem resistance & associated mortality in such infections.
Objectives
To screen for carbapenem resistance among gram negative infections in ICU patients by disc diffusion & confirmation by Vitek2 system.
To detect the carbapenemase production by phenotypic methods.
To confirm the presence of resistant gene by PCR.
To find out the attributable risk factors from the clinical history of such patients.
To follow up all the cases till the end of hospital stay & estimate mortality rate in such infections and its statistical correlation.
Materials and Methods
Source of data
Patients admitted in various Intensive care units of a tertiary care hospital, suspected of having Carbapenem resistant gram negative infections.
Study period
Study conducted over a period of one and half year from Jan- 2015 to July2016.
Study design
Cross sectional study.
Inclusion criteria
Patients of any age admitted in Intensive care units of a tertiary care hospital, suspected of having carbapenem resistant infections.
Patients from various Intensive care units whose samples yielded only gram negative bacteria.
Exclusion criteria
Patients not admitted in Intensive care units i.e admitted in general wards or treated in out patient department.
Patients from Intensive care units whose samples yielded gram positive bacteria.
Patients from Intensive care units whose samples yielded anaerobic bacteria.
Patients from Intensive care units whose samples were negative for bacterial culture.
All the relevant details of the patients included in the study, i.e name, age, sex, occupation, duration of illness, whether any invasive procedures or surgical intervention done, history of antibiotic usage, site of infection, past history, family history, were taken in a structured proforma.
Quality control
When performing phenotypic and genotypic tests, both positive and negative controls were used.
Positive control
Carbapenemase producing organism (Klebsiella pneumoniae ATCCBAA 1705).
Negative control
Non-carbapenemase-producing organism (Escherichia coli ATCC 25922).
Sample collection
A total of 160 non repetitive, clinical isolates of Gram negative bacteria from the patients admitted in various Intensive Care Units which were screened in the microbiology laboratory over a period of one and half year (Jan 2015 to Aug 2016), were identified, based on the colony morphology[3] and the biochemical reactions[4] from a variety of clinical samples like urine, stool, sputum, blood, pus were included in the study.
Screening test (Kirby bauer disk diffusion test): Isolates were screened for possible Carbapenemase production using Meropenem(10µg) disc. All the isolates showing meropenem disc diffusion zone diameter less than 21mm were considered to be screen positive.[5]
Phenotypic confirmatory methods
All the meropenem screen positive isolates were tested by the following phenotypic methods:
Vitek 2 system: All the resistant isolates were tested in Vitek 2 automated system (Biomeriux) by both ID and AST cards for accurate identification of the isolate and determining the MICs.
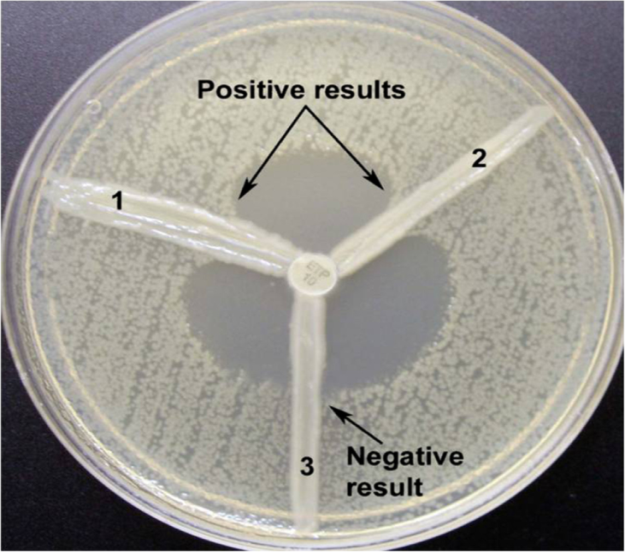
Modified Hodge test (MHT): Test isolates were subjected to Modified hodge test using E.coli ATCC 25922 as an indicator organism to detect the clover leaf zone of inhibition which signifies carbapenemase production.
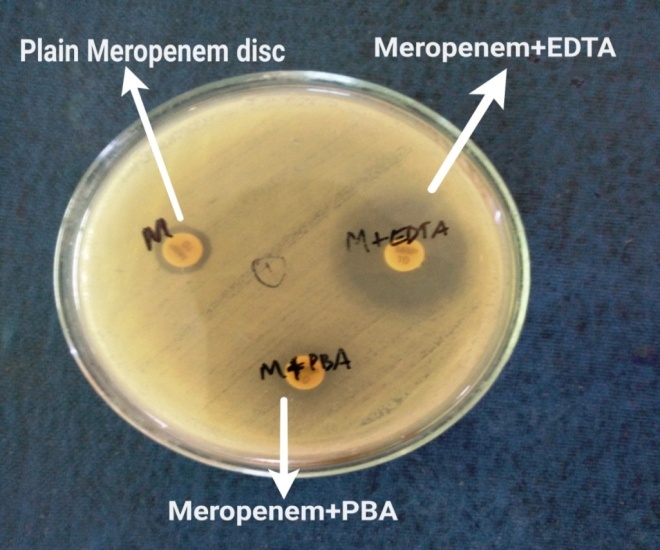
Combined disc test (CDT): [6] or detection of mrtallobetalactamase(MBL) production. Combined disc test was done using Muller Hinton agar with 10 µg plain meropenem disc and a meropenem disc with added inhibitors i.e, 20 micro litre of Phenyl boronic acid(PBA) (20mg/ml) for KPCs and 10 micro litre of 0.5M EDTA for metallobetalactamse production. The isolate was considered KPC producing when the growth inhibitory zone diameter around the meropenem disc with PBA increased ≥5mm when compared with the growth-inhibitory zone diameter around the disc containing meropenem alone. The isolate was considered MBL producing when the growth inhibitory zone diameter around the meropenem disc with EDTA and the meropenem disc increased ≥5mm when compared with the growth-inhibitory zone diameter around the disc containing meropenem alone.
Genotypic detection of carbapenemase genes
Multiplex PCR for detecting NDM, VIM, IMP and KPC genes:
The multiplex PCR was done according to the methodology described in the Sambrook and Russell editors. In Molecular Cloning: A Laboratory Manual.[7] Previously published primers were used by referring an article by Rachna Solanki et al., (2014)[8]
Primer sequences of 4 target genes (blaNDM-1, blaVIM, blaIMP and blaKPC genes) for Multiplex PCR: (Rachna Solanki et al., 2014) [Table 1].
|
Primer |
Orientation |
Oligonucleotide sequence(5΄-3΄) |
Size(bp) |
|
NDM-1 |
Forward |
GCATAAGTCGCAATCCCCG |
237 |
|
Reverse |
CTTCCTATCTCGACATGCCG |
||
|
VIM |
Forward |
GTTTGGTCGCATATCGCAAC |
382 |
|
Reverse |
AATGCGCAGCACCAGGATAG |
||
|
IMP |
Forward |
GAAGGCGTTTATGTTCATAC |
587 |
|
Reverse |
GTAAGTTTCAAGAGTGATGC |
||
|
KPC |
Forward |
TCGAACAGGACTTTGGCG |
201 |
|
Reverse |
GGAACCAGCGCATTTTTGC |
Results

Patients within the age group of 41-60yrs showed highest percentage of resistance.

Skin & soft tissue infections account for the highest resistance among various types of infection.


Surgical ICU showed the highest prevalence of carbapenem resistant among GNBs shown in [Figure 6].
Gentamicin(35%), Amikacin (45%), Tigecycline (88%) & Colistin(100%) were sensitive for carbapenem resistant GNBs. All the 3rd generation cephalosporins, Amoxicillin+clavulanic acid were 100% resistant to all carbapenem resistant gram negative bacilli shown in [Figure 7].

Significantly higher mortality was seen in carbapenem resistant infections.(p value 0.029) shown in [Figure 8].

Correlation between presence of co-morbidities as a risk factor for carbapenem resistance was highly significant.(p value 0.01) shown in [Figure 9].

Out of 20 deaths that occurred in ICU patients with carbapenem resistant infections, 13 cases (54%) were NDM-1 gene positive and 7 cases (24%) were genotype negative (p value is 0.026) i.e < 0.05 and hence presence of NDM-1 gene and mortality is statistically significant shown in [Figure 10].

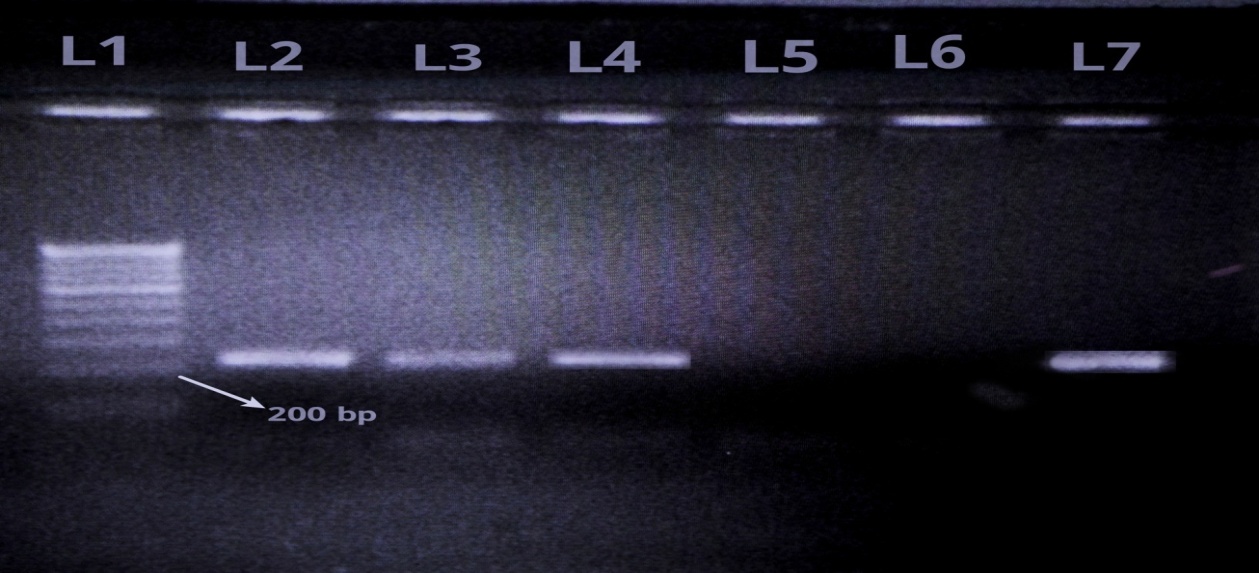
Of the total 160 gram negative isolates from ICU patients, 53 were resistant to carbapenems, Out of 53 resistant isolates, only 17 were MHT positive whereas 39 were CDT positive using EDTA as inhibitor and none of the isolate was positive using PBA as inhibitor. Multiplex PCR showed the presence of NDM-1 gene in 24 isolates whereas 29 isolates were negative for all the four genes.
|
Organism (n=160) |
Meropenem screen Positive by disc diffusion (n=53) |
MHT positive (n=17) |
Combined disc test positive (n=39) |
PCR positive (n=24) |
||||
|
with PBA for KPC |
with EDTA for MBL |
|||||||
|
NDM |
VIM |
IMP |
KPC |
|||||
|
E.coli (n=71) |
23 |
9 |
0 |
16 |
12 |
0 |
0 |
0 |
|
Klebsiella (n=63) |
17 |
5 |
0 |
13 |
7 |
0 |
0 |
0 |
|
Acinetobacter (n=10)) |
7 |
2 |
0 |
6 |
3 |
0 |
0 |
0 |
|
Pseudomonas(n=9) |
5 |
0 |
0 |
3 |
1 |
0 |
0 |
0 |
|
Enterobacter (n=7) |
1 |
1 |
0 |
1 |
1 |
0 |
0 |
0 |
|
S.No. |
Clinical characteristics |
Carabpenem sensitive Infections, (n=107) |
Carbapenem resistant Infections, (n=53) |
p value |
|
|
1 |
Mean Age |
45.4± 15 years |
41.49±14 years |
|
|
|
2 |
Gender |
Male |
58 |
34 |
0.028 |
|
Female |
49 |
19 |
0.23 |
||
|
3 |
Prior Hospitalization (last 3 months) |
16 |
15 |
0.045 |
|
|
4 |
Fever on admission |
13 |
3 |
0.19 |
|
|
5 |
ICU stay |
17.5 ± 2 days |
25.2 ± 7 days |
0.00016 |
|
|
6 |
Antibiotic exposure (last 3 months) |
23 |
31 |
0.000003 |
|
|
7 |
Invasive/surgical procedures in last 3 months |
25 |
22 |
0.018 |
|
|
8 |
Risk Factor |
Diabetes mellitus |
18 |
8 |
0.78 |
|
9 |
Coronary Artery disease |
9 |
6 |
0.55 |
|
|
10 |
Liver failure |
0 |
3 |
0.013 |
|
|
11 |
Respiratory illness |
4 |
3 |
0.5 |
|
|
12 |
Neurological illness |
2 |
1 |
0.4 |
|
|
13 |
Malignancies |
0 |
2 |
0.04 |
|
|
14 |
Mortality |
23(21%) |
20(37%) |
0.029 |
From the [Table 3], significant risk factors for development of carbapenem resistance in the present study are prior hospitalization, previous exposure to antibiotics, invasive procedures done in last 3 months, presence of co-morbidities and longer duration of ICU stay. Significantly higher mortality was seen in Carbapenem resistant cases(37%) compared to sensitive cases (21%).
Discussion
Carbapenem resistance can be due to presence of Carbapenemase enzymes, excessive production of ESBL or porin loss or efflux pumps or combination of more than one mechanism.
The prevalence of Carbapenem resistant gram negative bacilli among the patients admitted in ICUs in this study was found to be 33.12% which was closely correlating with Jamal Wafaa et al., 2016 (34.4%) from Kuwait and Kumari Seema et al.,2011(35%) from Varanasi. Studies from various parts of India had reported a varying prevalence ranging from as low as 14% by Chakrapani et al. from Chennai in 2014 to as high as 67% by Tempe DK et al. from New Delhi in 2015. Such a wide variation is probably due to variation in risk factors, extent of antibiotic use, variation in adherence to proper infection control practices, variation in knowledge of nursing staff regarding hospital infection control and lastly being a public sector or a private sector hospital to have proper infrastructural facilities for adequate patient care.
The highest prevalence of Carbapenem resistant gram negative infections in ICUs in the present study was observed in the patients of age group 41to 60 years which closely correlates with the findings of Vasundhara Devi et al., 2014 from Nellore and Humaira B et al.,2013 from Srinagar. Probably because this age group is more exposed to antibiotics, has more co-morbidities like diabetes mellitus, hypertension, coronary artery disease, etc for which they are more often hospitalized than the population of the other age groups.
In the present study, highest prevalence of carabpenem resistance was seen in patients of surgical ICU(23%) which correlated to the findings of Elandevi et al., 2016 (Chennai) who also reported highest prevalence among surgical department(47%).
Highest number of Carbapenem resistant Gram negative isolates was obtained from samples of patients suffering with Skin & soft tissue infections (51%), followed by UTI (24.5%). This closely correlates with Elandevi R et al., 2016 from Chennai which reported Carbapenem resistance in 38% surgical wound & 30% in UTI. In contrast, other studies from various places like Tempe DK et al.,2015 from New Delhi, Hengkoneng M et al., 2014 from Manipur, Humaira B et al., 2013 from Srinagar, PK Nair et al.[9] 2012 from Mumbai and RM Parveen et al.,[10] 2010 from Pondicherry reported highest cases of carbapenem resistant gram negative infection from Urinary tract infection.
The highest prevalence of carbapenem resistance in the present study was observed in Escherichia coli (43.4%) which correlates with Hengkoneng et al., 2014 (Manipur) and Sanjeev K et al.2015 (Udaipur).
Carbapenem resistant gram negative isolates in the present study showed maximum antimicrobial susceptibility to Colistin (100%), followed by Tigecycline (88%) which correlates with the findings of Chauhan K et al., (Meerut), 2015 who also reported Colistin (100%) & tigecycline (98%) sensitivity.
Present study showed that among phenotypic detection tests for carbapenemase production, Combined disc test is more sensitive than Modified Hodge test which is consistent with the findings of Sanjeev K et al., 2016 (Udaipur), Swetha et al.,2016(Mysuru) & Chauhan K et al., 2015 (Meerut).
NDM-1 gene was detected in 45% of carbapenem resistant isolates. Other genes like KPC, VIM, IMP were not detected in any isolate.
Contrary to this, studies from most of the European countries, USA & Canada reported a higher incidence of KPC gene responsible for carbapenem resistance Munoz-Price et al. in 2013 as reported by S Bratau et al., 2005 (USA) and Munoz-Price et al. in 2013 signifying endemicity of KPC
Mortality in Carbapenem resistant gram negative infections, in the present study was found to be 37.6% (p value<0.05) which correlated with Tian L et al.,2015 (China), Yang et al., 2015 (China) and Porwal et al., 2014 (Chennai)
58% of the patients who had carbapenem resistant infections were previously exposed to three or more antibiotics in the last three months (p value <0.05) which correlated with the findings of Ling M et al.,2015 (Singapore) 60%, K Kalam et al.,[11] 2014 (Pakistan) 62% and Chang HJ et al., 2010 (Taiwan) 52%.
Conclusion
Prevalence of carbapenem resistance is very high in Intensive care units, history of prior hospitalization should be considered before starting any antimicrobial therapy. Rapid detection tests like Combined disc tests can be used to detect metallobetalactamase production. Tigecycline, Colistin and to some extent Aminoglycosides can be used as therapeutic options for such infections and hence reduce morbidity & mortality.
Presence of co-morbidities, prior hospitalizations and previous exposure to antibiotics were significant risk factors for carbapenem resistant gram negative infections. NDM-1 genotype was associated with significant mortality among carbapenem resistant cases .Effective infection control measures should be taken to prevent nosocomial spread of these multidrug resistant pathogens.
Source of Funding
None.
Conflict of Interest
The authors declare that there is no conflict of interest.
References
- KM Papp-Wallace, A Endimiani, MA Taracila, RA Bonomo. Carbapenems: Past, Present, and Future. Antimicrob Agents Chemother 2011. [Google Scholar] [Crossref]
- HJ Morrill, JM Pogue, KS Kaye, KL LaPlante. Treatment Options for Carbapenem-Resistant Enterobacteriaceae Infections. Open Forum Infect Dis 2015. [Google Scholar] [Crossref]
- EW Koneman, PC Schreckenberger, SD Allen. . Konemans color atlas and textbook of diagnostic microbiology 2005. [Google Scholar]
- . Tests for Identification of Bacteria. Mackie & McCartney Practical Medical Microbiology 1996. [Google Scholar]
- . CLSI. Performance Standards for antimicrobial disc susceptibility tests. 11th ed., Approved Standards M 100 S 20, Clinical and Laboratory Standards Institute, Wayne. 2012. [Google Scholar]
- S Kumar, SK Mehra. Performance of Modified Hodge Test and Combined Disc Test for Detection of Carbapenemases in Clinical Isolates of Enterobacteriaceae Int. J Curr Microbiol App Sci 2015. [Google Scholar]
- Sambrook, Russell. . Molecular Cloning: A Laboratory Manual 2007. [Google Scholar]
- R Solanki, L Vanjari, S Subramanian, V Lakshmi. Comparative Evaluation of Multiplex PCR and Routine Laboratory Phenotypic Methods for detection of Carbapenemases among Gram Negative Bacilli. J Clin Diagn Res 2014. [Google Scholar]
- PK Nair. Prevalence of carbapenem resistant Enterobacteriaceae from a tertiary care hospital in Mumbai, India. J Microbiol Infect Dise 2013. [Google Scholar] [Crossref]
- R Mohamudha Parveen, B N Harish And, S C Parija. Emerging Carbapenem Resistance Among Nosocomial Isolates Of Klebsiella Pneumoniae In South India. International Journal of Pharma and Bio Sciences . [Google Scholar]
- K Kalam, F Qamar, S Kumar, S Ali, S Baqi. Risk factors for carbapenem resistant bacteraemia and mortality due to gram negative bacteraemia in a developing country. J Pak Med Assoc 2014. [Google Scholar]
- Introduction
- Objectives
- Materials and Methods
- Source of data
- Study period
- Study design
- Inclusion criteria
- Exclusion criteria
- Quality control
- Positive control
- Negative control
- Sample collection
- Phenotypic confirmatory methods
- Genotypic detection of carbapenemase genes
- Results
- Discussion
- Conclusion
- Source of Funding
- Conflict of Interest
