Introduction
Skin and soft tissue infections are a major healthcare issue worldwide and are of particular importance in developing countries.1 Factors that exacerbate skin infection cases in the developing countries are low level of hygiene, lack of access to clean water, malnutrition, and overcrowded living conditions in lower socio-economic classes.2
Pyogenic bacteria such as Staphylococcus aureus, Pseudomonas aeruginosa, Escherichia coli and streptococci are the prominent species involved in intricate multibacterial infections of deeper skin structures, major abscesses, burns and ulcers, bite wounds and diabetic.3 Even though most skin infections begin as local induration, erythema, pain or tenderness at the site of infection, the impact may be so worse that it can progress to life-threatening necrotizing fasciitis. When a healthy skin encounters a bacterial onslaught, epidermal cells (keratinocytes) stage robust innate immune response producing antimicrobial peptides and proinflammatory cytokines.4 However, with a progressing infection, pathogenic bacteria succeed in gaining entry to keratinocytes and evade host immune response. Since the majority of antibiotics cannot freely enter eukaryotic cells, cutaneous intracellular accumulation of pathogens like methicillin-resistant S. aureus (MRSA) poses a great challenge to treat skin infections using antibiotics.5 Further, the indiscriminate and reckless use of antibiotics has led to a crisis of multidrug resistance among bacterial pathogens. Infections caused by multidrug resistant microorganisms often fail to respond to conventional treatment, resulting in prolonged illness and greater risk of death. New resistance mechanisms have emerged in various bacteria, making the antibiotic therapy difficult.6 The longer duration of illness and treatment often in hospitals increases health-care costs and the economic burden to families and societies where it increases the costs of health care.7
During the recent past bacteriophage therapy has gained a renewed interest as a potential alternative therapy for multidrug-resistant (MDR) bacterial infections.8 Importantly, phages possess remarkable species specificity to bacterial pathogens. Further, an important clinical significance is that phage therapy can be very effective in biofilm infections where antibiotics fail to kill bacterial cells in biofilms, which are impermeable to antibiotics.9 Phage therapy finds special advantage for localized use, because phages penetrate deeper as long as the infection is present, rather than decrease rapidly in concentration below the surface. The phages stop reproducing once specific bacteria they target are destroyed. This prevents the development of secondary resistance to phages, which is quite often the case in antibiotics. Since phages live in the same niche as their host bacteria.10 and that they exhibit very high specificity (both species- and strain-specificity), it is crucial that phages are isolated and characterized for their bacteria-lytic ability locally in the affected regions. We recently demonstrated in an in vivo skin excisional wound model that the survival rate in mice was significantly high when a locally isolated phage against methicillin-resistant S. aureus was applied to the wounds.11 In this study we took up a systematic approach at first to isolate clinically important pathogenic bacterial species from pyogenic skin infections, determined antibiotic susceptibility of the isolates and then tested phages for lytic efficacy against MDR bacterial pathogen isolates.
Materials and Methods
Collection of clinical samples
The study was approved by the institutional Ethics Committee (Institutional Animal Ethics Committee-IAEC, Sree Siddaganga College of Pharmacy, Tumkur, Karnataka state, India-572 102 with an Approval reference No:SSCPT/IAEC.Clear/130/12-13). Written informed consent was obtained from all study patients (n=114). Swab samples from pyogenic skin infections were collected from patients of different age groups including males (n=82 median age 41 yrs) and females (n=32 median age 28 yrs). Swabs were immersed in 1% sterile peptone water and immediately subjected to microbial culture in aseptic conditions within 30-45 min of collection.
Isolation, identification and characterization of bacterial pathogens
The samples were at first cultured on general nutrient agar medium and suspect colonies were subsequently subcultured on selective agar media for different species. The major clinically important bacterial species from pyogenic infections were isolated using selective growth medium for each species. Identification of the species was achieved by extensive microbiological and biochemical characterization of presumptive colonies.
Antibiotic susceptibility of the bacterial isolates
The confirmed clinical isolates of each species were screened for antibiotic susceptibility by Kirby Bauer’s disc diffusion method.12, 13 according to Clinical and Laboratory Standard Institute (CLSI) guidelines (2005). Six hours old cultures were spread uniformly on the surface of Mueller-Hinton agar plates, antibiotic discs were placed on the agar surface and the plates were incubated at 37° C for 24 hr. The zone of inhibition was recorded in millimetres. The results were interpreted as susceptible or resistant as per the CLSI guidelines, 2005.14 Blank discs without antibiotic were used as negative controls in each experiment. The minimum inhibitory concentration (MIC) of methicillin was performed on S. aureus using the broth microdilution methods as described previously.15
DNA extraction, 16SrRNA gene amplification and phylogenetic analysis
Genomic DNA from the bacterial isolates was extracted by following a previously described method.16 Complete 16S rRNA gene from select strains from each species were amplified using the primers 16S-1-GGT GGA GCA TGT GGT TTA and 16S-2 (5'-16S-2 CCA TTG TAG CAC GTG TGT).17 PCR reaction mixture consisted of 200 µM dNTPS, 2.5 U Taq DNA polymerase and DNA template. After thermal cycling, the PCR products were gel purified using the QIAquick PCR Purification Kit. Following a nested PCR strategy,17 a 256-bp internal region of the amplified 16S rRNA gene was sequenced with an ABI 3100 automated sequencer (Applied Biosystems). The sequence of the PCR product was compared with known 16S rRNA gene sequences in the Gene Bank by multiple sequence alignment using the CLUSTAL W Program . Phylogenetic trees were constructed using Mega 5 program.
PCR amplification of mecA gene
To test whether the methicillin resistant S. aureus strains harbored a methicillin resistance gene (mecA), 4 of the MRSA isolates were subject to PCR by using the primers mecA-F (5'- CCT AGT AAA GCT CCG GAA) and mecA-R Primer (5'- CTA GTC CAT TCG GTC CA) as describe previously.18 The 25-μl reaction mixture included 200 μM dNTPs, 2.5 μM primers, 2.5 U of Taq DNA polymerase. Thermal profile was as follows: Initial denaturation for 5 min at 94°C, followed by 30 cycles of denaturation for 30s at 94°C; annealing for 30s at 53°C; and primer extension for 40 sec at 72°C and a final extension for 10 min at 72oC. A 10-µl PCR product was loaded onto a 1.2% agarose gel the bands were visualized under a UV transilluminator.
Isolation of bacteriophages
A cocktail of sewage originated from hospital, municipal and domestic waste in Davangere city, Karnataka, India, was used as a source for the isolation of phages. Initially, 25 ml of the cocktail sewage sample was pre-treated with 200 µl of chloroform for 15 minutes and to this, 2 ml of 16-18 hours old bacterial cultures, each species separately, were added and incubated overnight at 37°C. The lysate was centrifuged (4000 ×g, 10 min) and the supernatant was filtered through a 0.22 µm membrane filter and serially diluted using SM phage buffer. One hundred microliter from this stock was mixed with 100 µl of host bacterial suspensions. Bacterial cell suspension without phage filtrate was used as a negative control. After incubation (37 °C, 20 min), luria bertani agar (0.7%) was added to the above, mixed gently and poured on LB agar plates followed by incubation at 37°C for 15-16 hours. A few prominent and isolated plaques were recovered using a sterile gel cutter followed by suction. The bacteriophage particles were diffused out of agarose by adding chloroform. The phage preparations were stored at 4°C for further use.
Transmission electron microscopy
Transmission electron microscopy was carried out at the laboratory of Hans-W Ackermann, Professor Emeritus, Department of Microbiology and Immunology, Laval University, Quebec, Canada. A high titer bacteriophage lysate of 108 pfu ml-1 previously filtered through 0.22 µm Minisart Sartorius filter was centrifuged and treated with a fixative 1% glutaraldehyde. A drop of purified and fixed phage lysate was deposited on Formvar carbon-coated copper grids and negatively stained with 1% phosphotungstic acid (PTA). The grids were examined in Tecnai G2 Biotwin (Philips-Netherland) transmission electron microscope. The images of phages were captured and the sizes of head and tail were measured.
Determination of Phage host range (Spot test)
Bacterial isolates (4 from each species) were screened for phage lysis by a spot test. The suspensions of log-phase bacteria were added to 15 ml sterile tubes with molten 3 ml agarose maintained at 47°C. The contents were mixed and overlaid on pre-dried LB agar plates. The dried plates having bacterial layer, on which 5 µl of phage suspension (108 pfu ml-1) was dropped and incubated at 37°C without inverting the plates. The results were scored as a clear zone of complete lysis (++), partial lysis with turbidity (+) and no lysis (0).
Effect of storage period, pH, temperature and chloroform on the stability of the phages
The storage stability of the phages was determined at temperatures -40°C, -20°C, 4°C, and 20°C and also in presence of 20% (v/v) glycerol at -40°C and -20°C. Stability was also tested at pH 2, 4, 5, 6, 7, 8, 9 and 10. Phage titres were determined after 3, 6, 9 and 12 months of storage by double agar layer technique.
Statistics
Antibiotic resistance or susceptibility differences between the groups were determined using McNemar test. Differences in plaque titre values (pfu/ml) were determined using a non-parametric Mann-Whitney U test. A P value of < 0.05 was considered statistically significant. The statistical software SPSS version 25.0 (IBM Corp, Armonk, NY, USA) was used for all analyses.
Results
Bacterial pathogens from pyogenic skin infections
Among the 84 bacterial strains isolated from pyogenic skin infections of the study (Table 1), S. aureus was the most prevalent with 57 (67.85%) isolates followed by P. mirabilis 14 (16.66%), P. aeruginosa 7 (8.33%), E. coli 5 (5.95%) and S. xylosus 1 (1.19%). The highest prevalence of all bacteria (41, 48.80%) was recorded from abscess, followed by pyoderma (11, 13.09%), ulcers (9, 10.71%), cellulitis (5, 5.95%), wounds (5, 5.95%), diabetic foot (5, 5.95%), and post-operative infections (4, 4.76%). Only one S. aureus strain was isolated from burn wound infections.
Antibiotic susceptibility of the bacterial pathogens from pyogenic infections
All bacterial isolates (n=74) were subjected to antibiotic susceptibility testing (Table 2). S. aureus isolates were found to be resistant to most of the common antibiotics, with a highest (22/46, 48%) resistance for amikacin followed by 40% (18/46 strains) of the strains for ciprofloxacin. However, clindamycin and erythromycin were the only antibiotics to which significantly higher number of S. aureus isolates were susceptible (P<0.05). The single isolate from the S. xylosus species was sensitive to clindamycin and methicillin and resistant to all other antibiotics tested for Gram positive bacteria. From Gram negatives, significantly higher number of isolates were resistant to all the antibiotics tested except amikacin (P<0.05). P. aeruginosa isolates were resistant 100% (10 of 10 strains) for amoxicillin, 80% (8/10) for cephotaxime, 80% (8/10) for cefoperazone, 60% (6/10) and 40% for each of ceftazidime, ciprofloxacin, levofloxacin, tobramycin and amikacin respectively. E. coli isolates were resistant 50% (2/4) for amikacin and ceftazidime, 100% for cefoperazone (4/4), 75% (3/4) for each of the other antibiotics (Table 2).
Methicillin resistance among S. aureus strains
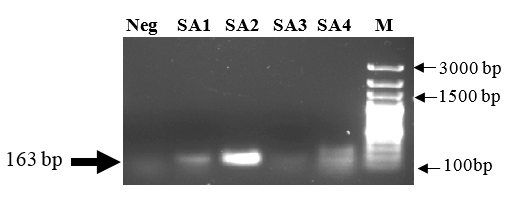
Considering the clinical significance of methicillin-resistant S. aureus strains, we also looked at the occurrence of MRSA strains among our S. aureus isolates. All strains of S. aureus having methicillin MIC of > 4 µg/ml were considered methicillin-resistant. As evidenced by antibiotic susceptibility tests, 41% of all S. aureus isolates were found to be methicillin-resistant (Table 2). When a molecular confirmation of methicillin resistance in these strains was sought, 3 strains showed an amplicon corresponding to the size of mecA gene (Figure 1). Similar to isolates from other species, these MDR S. aureus strains were selected for investigating the host range of the bacteriophage ΦDMSA-2.
Figure 1
Detection of mecA gene among methicillin resistant S. aureus isolates. Four of the multidrug resistant S. aureus strains (SA1-SA4) that were also resistant to methicillin were tested for the presence of mecA gene that encodes methicillin resistance. The amplicons were separated on a 2% agarose gel, stained with ethidium bromide and visualized under UV transilluminator.
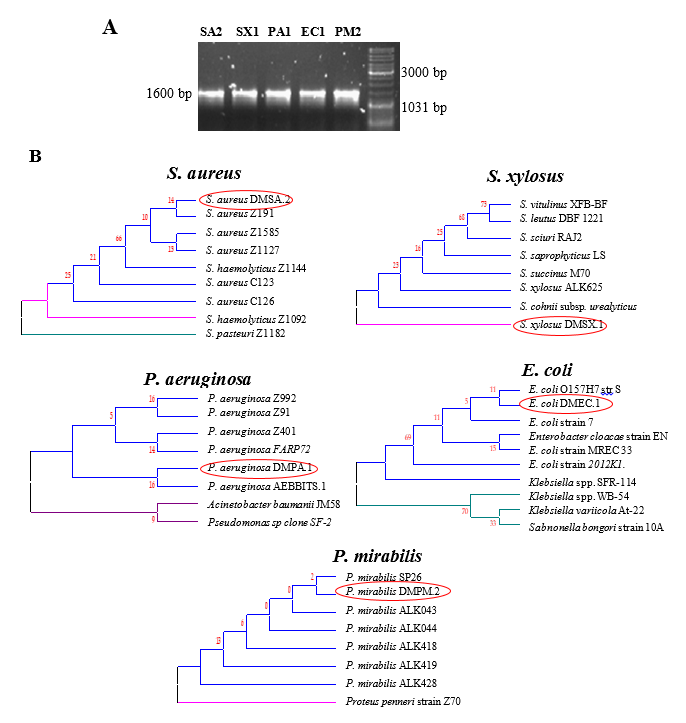
16S rRNA gene sequencing and phylogenetic analysis
Phylogenetic trees were constructed using 16S rRNA gene sequences from each of the MDR pathogens from this study against the sequences of MDR strains from NCBI (Figure 2 ). The phylogenetic trees revealed that the isolates formed a cluster with other MDR strains of the same species, but not with non-MDR strains or strains belonging to different species.
Figure 2
16S rRNA gene sequence based phylogenetic analysis of the MDR pathogen isolates. PCR amplification of the 16S rRNA gene from the representative strains from each pathogenic species (A). 16S rRNA gene sequences from the MDR bacterial isolates were compared with those from known MDR pathogens of the same species. Multiple sequence alignment and phylogenetic trees were constructed using the bioinformatics tool MEGA5 (B). Circled isolates are from this study
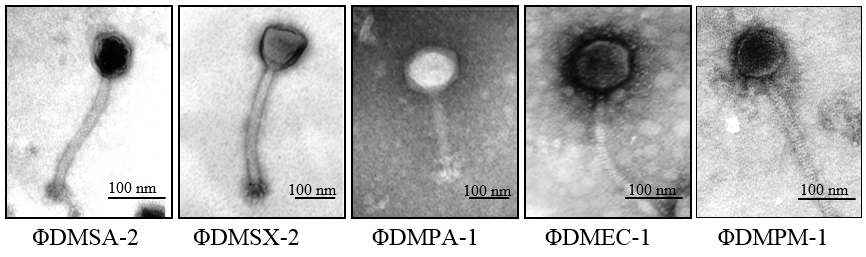
Isolation and morphological features of the phages
Figure 3 depicts transmission electron micrographs of all 5 phages, i.e., ΦDMSA-2, ΦDMSX-1, ΦDMPA-1, ΦDMEC-1, and ΦDMPM-1. All phages possessed an isometric icosahedral capsid of varying sizes. The details of phage dimensions and their classification are given in Figure 3. The phages ΦDMSA-2, ΦDMEC, and ΦDMPM exhibited a head size in the range 60-90 nm and tail of 200-240 nm long. These 3 phages were assessed to be belonging to the family Siphoviridae. The morphological characteristics of phage was examined and confirmed as phage belonging to the family Siphoviridae. On the other hand, phages ΦDMSX and ΦDMPA appeared to possess a bigger head size with 100-137 nm and a tail of 210-217 nm. Interestingly, the phage ΦDMSX for S. xylosus had a head with a triangular aspect, which was different from all other phages. ΦDMSX belongs to a group of "twort-like phages" under the sub family Spounavirinae of the family Myoviridae. Its tail measured about 210 nm showing conspicuous striations and terminate in a base plate with a set of spikes.
Figure 3
Electron micrographs of phages specific to the MDR pathogen isolates. Glutaraldehyde-fixed phage lysate was deposited on formvar carbon-coated copper grids and negatively stained with 1% phosphotungstic acid. TEM images were acquired on a Tecnai G Biotwin (Philips-Netherland) transmission electron microscope. Using the scale from the image acquiring software, sizes of head and tail were measured
Host range
Four multidrug-resistant strains from each species were chosen for determining the in vitro lytic efficacy of the phages ΦDMSA-2, ΦDMSX-1, ΦDMEC-1, ΦDMPA-1, and ΦDMPM-1 Table 3 shows the degree of lytic activity of all five phages against their specific host MDR isolates from each species. The results were scored as a clear zone of complete lysis (++), partial lysis with turbidity (+) and no lysis (0). The phages against S. aureus (ΦDMSA-2), P. aeruginosa (Φ DMPA-1) and E. coli (ΦDMEC-1) exhibited strong lytic activity and could completely lyse 3 of 4 (75%) tested strains. Importantly, no bacteriophage-insensitive mutants (BIM) were observed for this phage. The S. xylosus phage was able to completely lyse 1 strain (25%), partially lyse 2 strains (50%) and no lysis in the case of 1 strain (25%). The phage ΦDMPM-1 against P. mirabilis showed only partial lysis of all 4 strains tested. The occurrence of bacteriophage-insensitive mutants (BIM) based on the growth of bacterial colonies within the plaque were noticed in the case of three phages (Φ DMSX-1, Φ DMEC-1 and Φ DMPA-1).
Factors influencing the stability of the phages
The stability of hage lysates was tested in different storage conditions and period. The level of phage titre (108 pfu) did not decrease significantly (P<0.05) after 3 month storage at all of the tested temperatures, i.e., 6 months at -200C and 9 months at -400C with glycerol. However, at room temperature, the titer was reduced 30% from 108 pfu/ml to 7×107 pfu/ml. When phages were exposed to a high temperature of 60°C for 15 minutes, phage titers decreased significantly up to 106 pfu/ml (P<0.05). Phages were stable between pH range 6-9 and completely lysed at pH 2 and 4. Incubation at pH 4 and 37°C caused a 30% (7×107 pfu/ml) and 60% (4×107 pfu/ml) decrease in phage titre after 1 and 5 h of incubation, respectively.
Table 1
Profile of pathogenic bacterial isolates from pyogenic skin infections
Table 2
Antibiotic resistance of the bacterial pathogens from pyogenic infections
Table 3
Host range of the bacteriophages against MDR bacterial strains
|
Bacterial isolates |
Lytic activity of Phages |
BIM* |
|
|
Complete lysis (Figure 4) |
S. aureus |
Φ DMSA.2 |
|
|
DMSA.1 |
++ |
- |
|
|
DMSA.2 |
++ |
||
|
DMSA.3 |
++ |
||
|
DMSA.4 |
- |
||
|
|
|
|
|
|
S. xylosus |
Φ DMSX.1 |
|
|
|
DMSA.1 |
++ |
+ |
|
|
DMSA.2 |
+ |
||
|
DMSA.3 |
- |
||
|
DMSA.4 |
+ |
||
|
|
|
|
|
|
Bacteriophage-insensitive mutant colony (Figure 5) |
P. aeruginosa |
Φ DMPA.1 |
|
|
DMPA.1 |
++ |
+ |
|
|
DMPA.2 |
+ |
||
|
DMPA.3 |
++ |
||
|
DMPA.4 |
++ |
||
|
|
|
|
|
|
E. coli |
Φ DMEC.1 |
|
|
|
DMEC.1 |
++ |
+ |
|
|
DMEC.2 |
++ |
||
|
DMEC.3 |
- |
||
|
DMEC.4 |
++ |
||
|
|
|
|
|
|
Turbidity due to partial lysis; Several BIM colonies (Figure 6) |
P. mirabilis |
Φ DMPM.1 |
|
|
DMPM.1 |
+ |
|
|
|
DMPM.2 |
+ |
|
|
|
DMPM.3 |
+ |
|
|
|
DMPM.4 |
+ |
|
Discussion
In this study, locally isolated phages lysed multi drug resistant bacterial isolates belonging to important pathogenic species from skin infections. Clinical samples collected from pyogenic skin infections were collected from a total of 114 patients. The highest prevalence of bacteria, i.e., 41 (48.80%) was recorded from abscess, followed by pyoderma, ulcers, cellulitis, wounds and diabetic foot. Existing literature has revealed that the general microbiota of human skin infections includes S. aureus as a highly predominant species,19 but species from other genera such as Pseudomonas, Enterococcus, Escherichia, Enterobacter, Klebsiella., Proteus, Acinetobacter, and coagulase-negative Staphylococcus spp. are also commonly present.3
Nearly 40% of the S. aureus strains showed resistance to all major antibiotics available for Gram-positive bacteria during the period this study was conducted. The only other Gram-positive species, S. xylosus, was resistant to all tested antibiotics except clindamycin and methicillin. Among Gram-negative bacteria, 50-100% of all three species, P. aeruginosa, E. coli, and P. mirabilis, were resistant to all antibiotics tested except amikacin. Multidrug resistance among bacterial pathogens occurring in skin infections in India has been reported in a number of studies. These studies found that major pathogens such as S. aureus, E. coli, P. aeruginosa etc. were resistant (>60%) to major antibiotics including penicillin, erythromycin, co-trimoxazole, clindamycin, cefepime, and ciprofloxacin.20 Even though Gram-negatives are most frequently encountered, S. aureus is of immense clinical significance due to its role in skin infections. In our study, 41% of the S. aureus isolates were resistant to methicillin. MRSA have become a major challenge worldwide with varied prevalence depending on geographic region.21, 22 MRSA develop resistance through the acquisition of the mecA gene, which facilitates production of “penicillin-binding protein 2a” (PBP2a) that strengthens the cell wall and increases resistance to β-lactam antibiotics by blocking the β-lactam binding site.23 Penicillin-resistant strains of S. aureus predominate the nosocomial infections and currently only <5% of the S. aureus strains are penicillin-susceptible.24
Biological and geographical relationship among MDR strains is generally studied using 16S rRNA gene sequences and allows sequence comparison among strains from different geographic regions. In the present study, each of the representative MDR pathogen species formed a close clade in the phylogenetic tree with other MDR strains. Horizontal transfer of antibiotic resistance genes in the environment is a continuous natural phenomenon. Thus, phylogenetic analysis of MDR strains provides important information about their geographic distribution and may therefore help device better treatment strategies.25
Phages against S. aureus, P. aeruginosa and E. coli exhibited highest lytic activity as evidenced by spot test. The phage against P. mirabilis was able to lyse all 4 strains but only moderately and no complete lysis was observed for any strain. In earlier studies, phages have been reported to display broad host range, killing different strains from the same species, e.g., phage Stau2 could lyse 80% of the S. aureus isolates obtained from hospitals in Taiwan.13 In another study, E. coli pathogenic strains EHEC, EPEC, ETEC, and UPEC were effectively lysed by a phage.26 High selectivity and specificity of phages to their respective host bacteria provide a convenient yet effective way to eradicate bacterial pathogens both intracellular and otherwise in skin infections.
In our study, except for S. aureus, all other species produced bacteriophage insensitive mutants (BIMs). Even though phages have been successfully tested as alternative therapeutic agents in several human infections,27 emergence of BIMs is a serious concern. The bacterial resistance to phages can occur through different mechanisms, e.g., restriction modification, abortive infection, and more commonly through mutations in phage receptor sites on the bacterial cell surface, preventing them from attachment. However, bacteriophage insensitivity is known to come at a cost for the host bacteria, i.e., the BIM trait makes the bacteria compromise with their performance in other traits resulting in slower growth, decreased virulence, and even diminished resistance to various antibiotics.28
From the perspective of phage therapy in clinical medicine, stability of phages upon storage and implementation of appropriate storage conditions is critical for a successful outcome. Storage stability of all four phages examined under different parameters showed that there was no significant decrease in the phage titre except at room temperature. Interestingly, only ΦDMSA-2 was stable for 9 months, the longest duration of storage as compared to the other phages. There are a few studies reporting the use phage as in hand sanitizer solutions and as disinfectants to sanitize operating rooms and medical equipment.29 Further, a gel containing a cocktail of phages targeted nasal carriage of MRSA significantly reducing the incidence of MRSA transmission.30, 18 Thus, our data showing remarkable stability of the phages in a variety of conditions is an important step forward in developing phage therapy for skin infections.
One of the serious concerns about the use of phage therapy in vivo is a strong antibody response which would clear the phages more quickly.31 To circumvent this problem, phages with different antigenicity or with low immunogenicity could be prepared. Other drawbacks of phages as therapeutic agents are their often-narrow host ranges and the fact that phages are not always lytic under certain physiological conditions. Further, phage therapy triggers release of endotoxins from the target pathogen due to widespread bacterial cell lysis. This occurs also when antibiotics are used, but it could be countered with regulated use of phages.
Conclusions
Predominant occurrence of clinically important, multidrug resistant bacterial pathogens in skin infections highlights the severity of the problem. Multidrug resistance among the majority of the bacterial isolates underscores the existing global problem with the current antibiotic therapies. Phages have special advantage for localized use in skin infections, because, unlike antibiotics, they penetrate deeper in an infected tissue rather than decrease rapidly in concentration below the surface. Importantly, in our recent study,11 lytic efficacy of the phage ΦDMSA-2 against MRSA, was shown to significantly enhance the survival of mice in an in vivo skin excisional model. Thus, our study emphasizes the importance of need-based and locally isolated bacteriophages as potential alternative antimicrobial therapy.
Ethics
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institute and with the 1975 Helsinki declaration and its later amendments (revised in 2000) or comparable ethical standards. The study’s experimental design and protocol were approved by the Ethics Committee at Bapuji Institute of Engineering and Technology.
Author Contribution
MNS: Conception, design, experimentation, data analysis and manuscript writing. MK: Experimentation, data analysis and manuscript writing. DA: Data analysis, manuscript writing.
Source of Funding
The corresponding author is grateful to Vision Group on Science and Technology (VGST), Govt. of Karnataka Secretariat, India for the facilities created through funding in the department of Biotechnology for research under the schemes Seed Money to Young Scientist for Research (SMYSR) and Centers of Innovative Science and Engineering Education (CISEE).



