- Visibility 518 Views
- Downloads 62 Downloads
- Permissions
- DOI 10.18231/j.ijmr.2021.065
-
CrossMark
- Citation
Comparative study of manual conventional blood cultures versus automated blood culture system in cases of septicemia
Abstract
Introduction: Blood cultures are a proven gold standard method for the identification of causative agents of bloodstream infections. Identification of causative organism along with antibiotic susceptibility plays a pivotal role in proposing suitable antibiotic therapy. Automated blood culture systems show improved monitoring of blood cultures by reducing the time and by ensuring more accurate results when compared to the conventional blood culture system.
Aims and Objectives: To isolate the organism from given blood samples of a suspected case of septicemia and to compare the results of conventional and automated blood culture systems and to study the antimicrobial susceptibility pattern of the pathogens isolated.
Materials and Methods: A prospective study of 6 months period was conducted among 100 subjects attending the Department of Microbiology in a tertiary care hospital. Subjects with symptoms and signs of septicemia were included. 25ml of venous blood was drawn aseptically from the venipuncture site, of which 5ml of blood was inoculated into 50ml of Brain Heart Infusion bottle in conventional blood culture system and 10ml each into aerobic and anaerobic BACTEC PLUS bottle in Automated blood culture system BACTEC FX40.
Results: Overall, 48% and 60% of the samples revealed positive growth by the conventional and automated blood culture system BACTEC FX40, respectively. Gram Positive Cocci were 52.08% and Gram Negative Bacilli were 47.91% isolated by conventional blood culture system, whereas automated blood culture system BACTEC FX40 isolated 45% and 55%, respectively. Isolates were detected within 24-48hrs and 12-24 hrs by conventional and automated blood culture systems, respectively. The anti-microbial susceptibility pattern of the pathogens isolated was also recorded by Kirby Bauer disc diffusion method of antimicrobial susceptiblity testing.
Conclusion: Automated blood culture systems are a trustworthy substitute to conventional blood culture systems. The automated blood culture systems being more sensitive and rapid in detecting septicemia in subjects acts as an appropriate means for the initial identification and detection of blood pathogens and improved provision of antimicrobial therapeutic options for septic Patients especially in Critical Care and Intensive Care Units where positive culture reporting is crucial.
Introduction
Septicemia or sepsis results when circulating bacteria in blood multiply at a rate that surpasses their elimination by phagocytes.[1] Blood infections are a substantial reason for morbidity and mortality of patients, particularly in developing countries.[2] If left untreated, bloodstream infections may lead to more dangerous infections, involving all organs and ultimately death.[3] Among the various types of nosocomial infections, bloodstream infections are a very serious health problem in hospital wards globally.[4]
Laboratory blood cultures are a proven standard tool for the identification of causative agents of bloodstream infections.[5] Blood cultures provide us information on the causative organism and their antibiotic susceptibility.[6]
This leads to a need for the most effective use of all the accessible procedures for the initial identification of microorganisms causing blood stream infections, which comprises conventional and automated blood culture systems. Technological developments resulted in the accessibility of diverse systems, each appealing to be greater in different facets.[7] Drawbacks of the conventional method require a better diagnostic tool with higher yield and speed.
An automated Blood Culture System is a new ray of hope in the diagnosis of bloodstream infection as it monitors continuously with higher sensitivity, specificity, and faster turnaround time.[8] With this background, the study was undertaken to compare the bacteriological outline and antibiotic susceptibility pattern by both conventional blood culture systems and automated BACTEC FX40 blood culture systems in cases of septicaemia.
Materials and Methods
A prospective study of six months period was conducted among 100 subjects attending the Microbiology department of Shadan Institute of Medical Sciences. Subjects with symptoms and signs of septicemia were included in the study.
Inclusion criteria
All the cases with the signs of septicemia like fever, chills, malaise, tachycardia, hyperventilation, and toxiciity or prostration were included.
Exclusion criteria
Cases without symptoms and signs of septicemia were not taken into account.
Blood culture by conventional blood culture system
As per standard procedures described in Bailey & Scott 12th edition,[9] 5ml of blood was drawn aseptically into a 50ml bottle of Brain Heart Infusion bottle with 0.05% Sodium polyanethol Sulfonate and were incubated at 37º C under aerobic conditions. After overnight incubation, (12-18hrs) Gram’s stain was done from the broth.
Blind subculture onto Blood agar, Chocolate agar, and MacConkey agar was done with help of a loop. Blood agar and MacConkey agar plates were incubated at 37º C under aerobic conditions overnight. Chocolate agar plate was incubated in a candle jar with 5-10% CO2 at 37º C overnight. All the plates were examined for growth and colony characteristics.
Subcultures were done into appropriate liquid media. Preliminary tests like Gram’s stain, hanging drop, catalase, oxidase test were done and appropriate biochemical reactions like Slide Coagulase, Tube Coagulase, Bile Aesculin Agar, Optochin Sensitivity, Bacitracin Sensitivity, DNAse test were put for Gram Positive Cocci and Indole Test, Methyl Red Reaction, Voges –Proskaeur test, Citrate and Urease Reaction, Triple Sugar Iron Agar, Nitrate Reduction Test and Decarboxylases Test for Gram Negative Bacilli. Culture negative bottles were subcultured again after 48hrs, 72hrs, and finally after 7 days of incubation. Bottles were inspected daily for any macroscopic evidence of growth and subcultured if turbidity, gas formation, lysis of blood was appreciated.
Blood culture by automated blood culture system
For the automated blood culture system, 10ml each of blood was inoculated into Aerobic and Anaerobic BACTEC PLUS bottles and were loaded into the machine. As soon as an audible or visible alert was given by the BACTEC FX40, it was treated as a positive blood culture and processed in a manner similar to conventional blood culture system by subculturing. If there was no alert, audible or visible, the bottles were incubated for 7 days before undergoing a terminal subculture, to report as negative.
Antibiotic susceptibility testing
Antibiotic susceptibility testing was carried out on Mueller Hinton agar using Kirby Bauer Disc Diffusion method[10] as per NCCLS guidelines by using the Hi Media Antibiotic Discs.
Statistical analysis of data
It was performed utilizing the SPSS software. Data analysis was done by, descriptive analysis, student t test and chi-square test, p value <0.05 was considered as significant.
Results
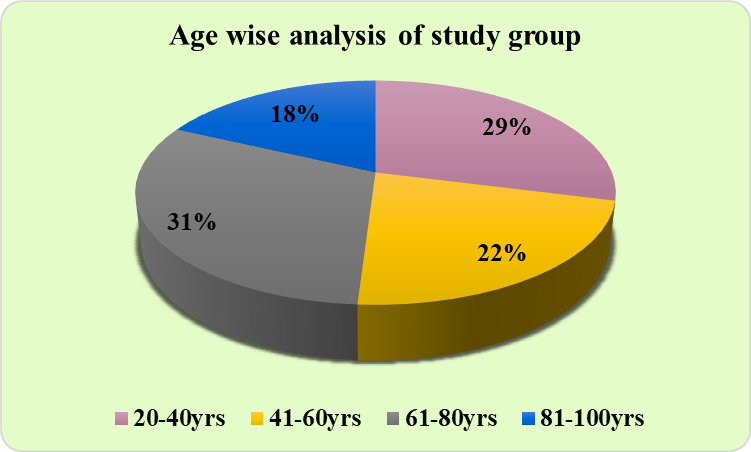
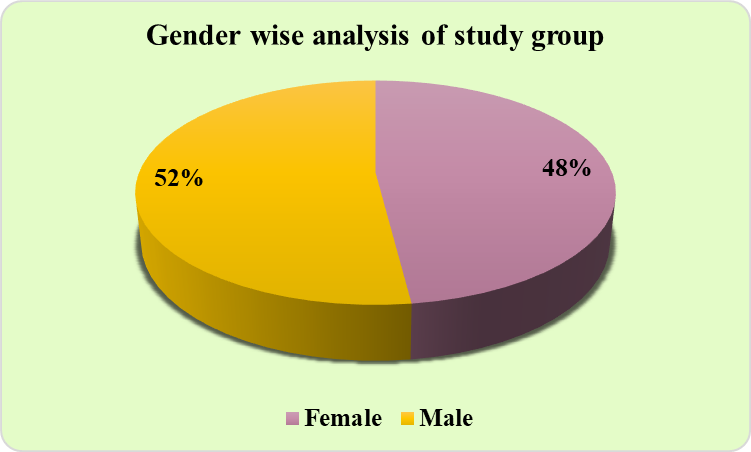
A total of 100 subjects diagnosed with septicemia participated, ranging from age 20 to 100 years, the majority of the subjects belonged to the 61-80 years age group (31%). ([Figure 1]). Male predominance (52%) was observed, and the females were 49% in our study. ([Figure 2]).
Blood was collected (5ml) and was inoculated into 50ml of Brain Heart Infusion bottle by using conventional blood culture system and 10ml each into Aerobic and Anaerobic BACTEC PLUS bottle by using automated blood culture system, for the identification of the microorganism and antibiotic sensitivity.
Out of 100 cases of clinically suspected septicemia, culture positive cases were 48. The isolation percentage being 48%. The various isolates were
Gram Positive cocci- 25 (52.08%)
Gram Negative bacilli – 23(47.91%)
The predominant organism was Coagulase Negative Staphylococcus followed by Escherichia coli. ([Table 1])
Out of 100 cases of clinically diagnosed septicemia, 60 cases were culture positive isolated by BACTEC FX40. The percentage of isolation being 60%. The various organisms isolated were
Gram positive cocci- 27(45%)
Gram negative bacilli – 33(55%)
The predominant organism found was – Coagulase Negative Staphylococcus. No anaerobic organisms were isolated. ([Table 2]).
Sensitivity patterns of Gram positive and Gram Negative isolates:
When sensitivity pattern of Gram positive isolates was studied, Coagulase Negative staphylococci showed 100% sensitivity to Vancomycin, Linezolid and Tetracycline followed by Trimethoprim-Sulfamethoxazole (83.3%), Clindamycin (58.3%), Oxacillin (50%), and Erythromycin (25%).
Staphylococcus aureus showed 88.9% sensitivity to Linezolid, 77.8% sensitivity to Vancomycin and Tetracycline, followed by Clindamycin (66.7%), Erythromycin (44.5%), and 33.4% to Oxacillin and Trimethoprim- Sulfamethoxazole.
Enterococcus showed 100% sensitivity to Vancomycin, Linezolid and Ampicillin.
Streptococcus showed 100% sensitivity to Vancomycin, followed by 75% sensitivity to Ampicillin, Ciprofloxacin and Erythromycin. 50% sensitivity to Penicillin, Tetracycline and Trimethoprim- Sulfamethoxazole. ([Table 3])
Amongst Gram Negative Bacilli, Enterobacter showed 100% sensitivity to Amikacin, Meropenem, and Ciprofloxacin.
Escherichia coli showed 100% sensitivity to Amikacin followed by Meropenem and Imipenem, Piperacillin- Tazobactam (90.9%) followed by Gentamycin (81.8%), then Ciprofloxacin and Trimethoprim- Sulfamethoxazole (36.4%).
Klebsiella showed 75% sensitivity to Amikacin, Ciprofloxacin, Gentamicin, Trimethoprim-Sulfamethoxazole, 62.5% sensitivity to Amoxyclav, 50% sensitivity to Levofloxacin and Piperacillin-Tazobactam, and 25% sensitivity to Cefepime, Imipenem, and Meropenem.
Proteus showed 100% sensitivity to Amikacin, Cefepime, Ciprofloxacin, Gentamicin, Levofloxacin, Meropenem, Amoxyclav, Piperacillin- Tazobactam and Trimethoprim-Sulfamethoxazole.
Acinetobacter showed 100% sensitivity to Amikacin, Cefepime, and Piperacillin - Tazobactam.
Alcaligenes fecalis showed 100% sensitivity to Amikacin, Ciprofloxacin, Levofloxacin, Trimethoprim- Sulfamethoxazole. Burkholderia- its strains showed 100% sensitivity to Levofloxacin and Trimethoprim-Sulfamethoxazole.
Pseudomonas showed 100% sensitivity to Amikacin, Gentamycin and Meropenem. 66.7% sensitivity to Amoxyclav. 50% sensitivity to Piperacillin- Tazobactam and Trimethoprim- Sulfamethoxazole. 33.4% sensitivity to Ciprofloxacin, Levofloxacin, Cefepime, Aztreonam. ([Table 4])
|
S. No |
Organism |
No. isolated |
Percentage of isolation |
|
1 |
Coagulase Negative Staphylococcus |
12 |
25% |
|
2 |
Enterococcus |
01 |
2.08% |
|
3 |
Staphylococcus aureus |
09 |
18.75% |
|
4 |
Streptococcus pneumoniae |
03 |
6.25% |
|
5 |
Klebsiella spp |
06 |
12.5% |
|
6 |
Escherichia coli |
10 |
20.84% |
|
7 |
Pseudomonas aeruginosa |
04 |
8.33% |
|
8 |
Proteus |
03 |
6.25% |
|
Total |
48 |
100% |
|
S. No |
Organism |
No. isolated |
Percentage of isolation |
|
1 |
Coagulase Negative Staphylococcus |
12 |
20% |
|
2 |
Staphylococcus aureus |
10 |
16.67% |
|
3 |
Enterococcus |
01 |
1.67% |
|
4 |
Streptococcus |
04 |
6.67% |
|
5 |
Enterobacter |
01 |
1.67% |
|
6 |
Escherichia coli |
11 |
18.33% |
|
7 |
Klebsiella |
08 |
13.33% |
|
8 |
Morganella |
01 |
1.67% |
|
9 |
Proteus |
03 |
5% |
|
10 |
Acinetobacter |
01 |
1.67% |
|
11 |
Alcaligenes fecalis |
01 |
1.67% |
|
12 |
Burkholderia |
01 |
1.67% |
|
13 |
Pseudomonas aeruginosa |
06 |
10% |
|
Total |
60 |
100% |
|
Organisms |
AMP |
CD |
CP |
E |
LZ |
OX |
P |
TE |
COT |
VA |
|
CONS |
- |
58.3% |
- |
25% |
100% |
50% |
- |
100% |
83.3% |
100% |
|
Staphylococcus aureus |
- |
66.7% |
- |
44.5% |
88.9% |
33.4% |
- |
77.9% |
- |
77.9% |
|
Enterococcus |
100% |
- |
- |
- |
100% |
- |
- |
- |
- |
100% |
|
Streptococcus pneumoniae |
75% |
- |
75% |
75% |
- |
- |
50% |
50% |
50% |
100% |
|
Organisms |
AMK |
AMP |
AO |
CPM |
CP |
G |
I |
LE |
MRP |
AU |
PT |
COT |
|
Enterobacter |
100% |
- |
- |
- |
100% |
- |
- |
- |
100% |
- |
- |
- |
|
Escherichia coli |
100% |
- |
- |
- |
36.4% |
81.8% |
100% |
- |
100% |
- |
90.9% |
36.4% |
|
Klebsiella |
75% |
- |
- |
25% |
75% |
75% |
- |
50% |
|
62.5% |
50% |
75% |
|
Proteus spp |
100% |
- |
- |
- |
100% |
100% |
100% |
- |
100% |
- |
100% |
100% |
|
Acinetobacter spp |
100% |
- |
- |
- |
100% |
- |
- |
- |
- |
- |
100% |
- |
|
Alcaligenes spp |
100% |
- |
- |
- |
100% |
- |
- |
100% |
- |
- |
- |
100% |
|
Burkholderia spp |
- |
- |
- |
- |
- |
- |
- |
100% |
- |
- |
- |
100% |
|
Pseudomonas aeruginosa |
100% |
- |
33.4% |
33.4% |
33.4% |
100% |
- |
33.4% |
100% |
66.7% |
50% |
50% |
Discussion
Detection of septicemia in the initial stages plays a crucial role in helping the diagnosis and management of suspected cases. Morbidity and mortality are closely associated with septicemia. On-time recognition of microorganisms can have a positive effect on the ultimate outcome as the microbial isolation from blood has a great diagnostic and prognostic implication and purpose and its antibiogram can clearly guide the clinician about an accurate regimen for the treatment.
Our study aimed at identifying isolates causing bloodstream infections in critical care units and wards and their antibiotic susceptibility. A comparative study was carried out between conventional and automated blood culture systems.
In the automated blood culture, out of 100 clinical cases of septicemia, 60 isolates were isolated i.e., the rate of positive blood culture was 60% whereas in the conventional blood culture system only 48 isolates were isolated. In the study conducted by Azra S Hasan et al, the rate of positive blood culture was 45.5% with Automated Blood Culture System[11] which correlates with the culture positivity rate which is 60% in our case.
In the BACTEC system, out of 100 cases, the percentage of isolation was 60%, Gram positive cocci accounted for 27 (45%). The most common pathogen being Coagulase negative Staphylococcus, followed by Escherichia coli. This study correlated with the study conducted by Sarangi et al in 2016[12] and with the study conducted by Qursheed Sultana, Humera Ansari et al in 2016 where a predominance of Gram positive isolates like Staphylococcus aureus & Coagulase negative Staphylococci was observed.[13]
Amongst Gram positive Cocci, Coagulase Negative staphylococci showed 100% sensitivity to Vancomycin followed by Linezolid and Tetracycline. This is correlated with the study conducted by Shahsanam Gheibi et al, where extreme sensitivity was discovered to Vancomycin.[14] Staphylococcus aureus showed major (88.9%) sensitivity to Linezolid, followed by Vancomycin and Tetracycline. It is associated with the study done by Jones RN et al in 2006.[15] Enterococcus showed 100% sensitivity to Vancomycin, Linezolid, and Ampicillin. It was relatable to the study done by Yadav G et al,[16] where sensitivity was observed with Vancomycin and Linezolid. Streptococcus showed 100% sensitivity to Vancomycin, followed by sensitivity to Ampicillin, Ciprofloxacin, and Erythromycin.[17]
Amongst Gram Negative Bacilli, Enterobacter showed 100% sensitivity to Amikacin, Meropenem, and Ciprofloxacin. Garcinuño P et al, in their study, showed a higher microbiological success rate with Amikacin and Meropenem.[18] Escherichia coli showed 100% sensitivity to Amikacin followed by Meropenem and Imipenem. A study done by Kidwai S et al showed Escherichia coli with highest sensitivity to Imipenem followed by Amikacin.[19] Klebsiella,[20] Proteus,[21] Acinetobacter[22] and Pseudomonas[23] showed maximum sensitivity to Amikacin. Burkholderia strains showed 100% sensitivity to Levofloxacin[24] and Trimethoprim-Sulfamethoxazole.[25]
The automated system showed 60% positivity as compared to 48% by conventional blood culture system of bacterial pathogens. Automated blood culture provides improved therapeutic results by enhancing the speed of the blood culture report within 12-16 hrs and the percentage of positivity. The conventional system is cost effective but, the automated blood culture system is more sensitive and rapid in detecting septicemia in patients.
Automated blood culture systems are a trustworthy substitute to conventional blood culture systems. The automated blood culture systems being more sensitive and rapid in detecting septicemia in subjects acts as an appropriate means for the initial identification and detection of blood pathogens.
Source of Funding
None.
Conflict of Interest
None.
References
- Koneman E, Allen S. . Koneman. Diagnostico Microbiologico/ Microbiological diagnosis: Texto Y Atlas En Color/ Text and Color Atlas. 2008. [Google Scholar]
- Deku J, Dakorah M, Lokpo S, Orish V, Ussher F, Kpene G. The Epidemiology of Bloodstream Infections and Antimicrobial Susceptibility Patterns: A Nine-Year Retrospective Study at St. Dominic Hospital. J Trop Med. 2019. [Google Scholar] [Crossref]
- Alizadeh A, Movahed R, Mohammadnia M. Comparative evaluation of conventional and bactec methods for detection of bacterial infection. Tanaffos. 2016;15(2):112-6. [Google Scholar]
- Khan H, Baig F, Mehboob R. Nosocomial infections: Epidemiology, prevention, control and surveillance. Asian Pac J Trop Biomed. 2017;7(5):478-82. [Google Scholar]
- Lamy B, Dargère S, Arendrup M, Parienti J, Tattevin P. How to optimize the use of blood cultures for the diagnosis of bloodstream infections? A state-of-the art. Front Microbiol. 2016;7. [Google Scholar] [Crossref]
- Prakash K, Arora V, Geethanjali P. Bloodstream bacterial pathogens and their antibiotic resistance pattern in Dhahira region Oman. Oman Med J. 2011;26(4):240-79. [Google Scholar]
- Rajan L, Jayalekha B, Sreekumary P, Harikumar S. A comparative study on conventional and automated blood culture in the early detection of bacterial pathogens. J Evol Med Dent Sci. 2017;6(31). [Google Scholar]
- Minassian A, Newnham R, Kalimeris E, Bejon P, Atkins B, Bowler I. Use of an automated blood culture system (BD BACTECTM) for diagnosis of prosthetic joint infections: Easy and fast. BMC Infect Dis. 2014;14(1). [Google Scholar]
- Forbes B, Sahm D, Weissfeld A. . Bailey and scott s diagnostic microbiology. 2007;53. [Google Scholar]
- Hudzicki J. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol Author Information. American Society For Microbiology. . 2009. [Google Scholar]
- Hasan A, Uppal P, Arya S, Capoor M, Nair D, Chellani H. Comparison of BacT/Alert microbial detection system with conventional blood culture method in neonatal sepsis. J Pediatr Infect Dis. 2008;3(1):21-5. [Google Scholar]
- Sarangi K, Pattnaik D, Mishra S, Nayak M, Jena J. Bacteriological profile and antibiogram of blood culture isolates done by automated culture and sensitivity method in a neonatal intensive care unit in a tertiary care hospital in Odisha, India. Int J Adv Med. 2015;2(4):387-92. [Google Scholar] [Crossref]
- Sultana Q, Ansari H, Ansari M. Bacteriological profile and antimicrobial susceptibility patterns of organisms responsible for blood stream infections. Indian J Microbiol Res. 2016;3(2):113-7. [Google Scholar]
- Gheibi S, Fakoor Z, Karamyyar M, Khashabi J, Ilkhanizadeh B, Asghari-Sana F. Coagulase negative staphylococcus; the most common cause of neonatal septicemia in Urmia, Iran. Iran J Pediatr. 2008;18(3):237-43. [Google Scholar]
- Jones R, Ross J, Fritsche T, Sader H. Oxazolidinone susceptibility patterns in 2004: Report from the Zyvox® Annual Appraisal of Potency and Spectrum (ZAAPS) Program assessing isolates from 16 nations. J Antimicrob Chemother. 2006;57(2):279-87. [Google Scholar]
- Yadav G, Thakuria B, Madan M, Agwan V, Pandey A. Linezolid and vancomycin resistant enterococci: A therapeutic problem. J Clin Diagn Res. 2017;11(8):7-11. [Google Scholar]
- Tuohy M, Washington J. Antimicrobial susceptibility of viridans group streptococci. Diagn Microbiol Infect Dis. 1997;29(4):140-5. [Google Scholar]
- Garcinuño P, Santibañez M, Gimeno L, Sánchez-Bautista A, Coy J, Sánchez-Paya J. Empirical monotherapy with meropenem or combination therapy: the microbiological point of view. Eur J Clin Microbiol Infect Dis. 2016;35(11):1851-55. [Google Scholar]
- Kidwai S, Nageen A, Ghaznavi S, Bashir F, Ara J. Antibiotic susceptibility in commonly isolated pathogens from urinary tract infection in a cohort of subjects from low socioeconomic strata. Pak J Med Sci. 2017;33(2):254-9. [Google Scholar]
- Patilaya P, Husori D, Marhafanny L. Susceptibility of klebsiella pneumoniae isolated from pus specimens of post-surgery patients in Medan, Indonesia to selected antibiotics. Open Access Maced J Med Sci. 2019;7(22):3861-4. [Google Scholar] [Crossref]
- Lazarević G, Petreska D, Pavlović S. Antibiotic sensitivity of bacteria isolated from the urine of children with urinary tract infections from 1986 to 1995. Srp Arh Celok Lek. 1998;126(11-12):423-9. [Google Scholar]
- Jung S, Yu J, Shin S, Park K, Jekarl D, Han K. Brief communication: False susceptibility to amikacin by VITEK 2 in acinetobacter baumannii harboring armA. Ann Clin Lab Sci. 2010;40(2):167-71. [Google Scholar]
- Javiya V, Ghatak S, Patel K, Patel J. Antibiotic susceptibility patterns of Pseudomonas aeruginosa at a tertiary care hospital in Gujarat, India. Indian J Pharmacol. 2008;40(5):230-4. [Google Scholar]
- Sethi S, Sharma M, Kumar S, Singhal L, Gautam V, Ray P. Antimicrobial susceptibility pattern of Burkholderia cepacia complex & Stenotrophomonas maltophilia from North India: Trend over a decade. Indian J Med Res. 2007;152(6):656-61. [Google Scholar]
- Dance D, Davong V, Soeng S, Phetsouvanh R, Newton P, Turner P. Trimethoprim/sulfamethoxazole resistance in Burkholderia pseudomallei. Int J Antimicrob Agents. 2014;44(4):368-9. [Google Scholar]
- Abstract
- Introduction
- Materials and Methods
- Inclusion criteria
- Exclusion criteria
- Blood culture by conventional blood culture system
- Blood culture by automated blood culture system
- Antibiotic susceptibility testing
- Statistical analysis of data
- Results
- Discussion
- Source of Funding
- Conflict of Interest
- References
How to Cite This Article
Vancouver
Saher L, Afzal M, Ansari HQF. Comparative study of manual conventional blood cultures versus automated blood culture system in cases of septicemia [Internet]. Indian J Microbiol Res. 2021 [cited 2025 Sep 22];8(4):327-332. Available from: https://doi.org/10.18231/j.ijmr.2021.065
APA
Saher, L., Afzal, M., Ansari, H. Q. F. (2021). Comparative study of manual conventional blood cultures versus automated blood culture system in cases of septicemia. Indian J Microbiol Res, 8(4), 327-332. https://doi.org/10.18231/j.ijmr.2021.065
MLA
Saher, Lubna, Afzal, Mustafa, Ansari, Humera Qudsia Fatima. "Comparative study of manual conventional blood cultures versus automated blood culture system in cases of septicemia." Indian J Microbiol Res, vol. 8, no. 4, 2021, pp. 327-332. https://doi.org/10.18231/j.ijmr.2021.065
Chicago
Saher, L., Afzal, M., Ansari, H. Q. F.. "Comparative study of manual conventional blood cultures versus automated blood culture system in cases of septicemia." Indian J Microbiol Res 8, no. 4 (2021): 327-332. https://doi.org/10.18231/j.ijmr.2021.065
