- Visibility 67 Views
- Downloads 11 Downloads
- DOI 10.18231/j.ijmr.2022.044
-
CrossMark
- Citation
Emergence of SARS-CoV-2 variant of concern omicron: biological features and genomic concern
Introduction
On 26th November, the World Health Organization (WHO)’s Technical Advisory Group on SARS-CoV-2 Virus Evolution (TAGSE) declared the Phylogenetic Assignment of Named Global Outbreak Lineages (PANGO) lineage B.1.1.529 as a VOC and designated it with the Greek letter Omicron.[1], [2], [3] The number of mutations that this variant has undergone makes it so concerning that these are some of the most frequently asked questions these days such as; will this variant be more transmissible, as the world has speculated? Will the symptoms become more severe? Do our immunizations still protect us? The B.1.1.529 variant encodes 37 amino acid changes in the spike (S) protein. Out of 37 gene mutations, 15 mutations are in the receptor-binding domain (RBD), which raises questions about the efficacy of current vaccinations and antibody treatments.[4] The mutations to the spike protein are the interest of concern (IOC) because if the spike protein changes enough or in certain key places, it might make our vaccines less effective. This variation further has led to affect the transmissibility, immune system evasion, and vaccine resistance. So far, five VOC namely Alpha, Beta, Gamma, Delta and Omicron variants, and all had around 8-15 mutations to the spike protein.[5] However, 15 mutations itself reported in the RBD of the spike protein of Omicron.[4] Hence the Omicron variant has raised the alarm because the mutations in the new variant could make it more transmissible than previous variants and already been reported in nearly 115 countries.[6], [7] Therefore, WHO quickly designated Omicron as a VOC. Headache, cough, shortness of breath, fever, fatigue, sore throat, muscle or body aches, and loss of taste or smell are the commonly observed symptoms of disease caused by the Omicron variant.[6] The reverse transcriptase–polymerase chain reaction (RT-PCR) is the gold standard and widely accepted method for the SARS-CoV-2 variant, including Omicron.[3], [6] To verify the presence of the virus, the approach used to look for specific gene amplification such as Spike (S), Enveloped (E), and Nucleocapsid (N). However, since the S gene has been highly altered in Omicron, many available primers may produce results indicating the absence of the A gene (S gene dropout). In combination with the detection of other viral genes, this S gene dropout could be utilized to diagnose Omicron. However, genomic sequencing is necessary to confirm the Omicron variant.
Evolution of SARS-CoV-2 and its sub-variants
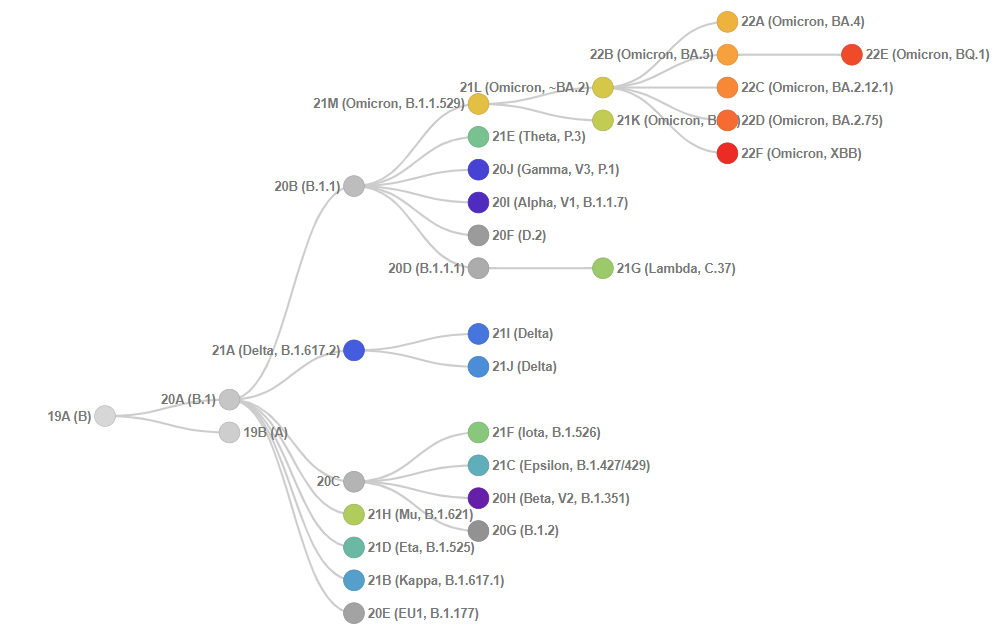
Genetic diversity, selected through evolutionary pressures, drives Corona virus (CoV) co-evolution with their hosts. SARS-CoV-2 genetic diversity is enabled by higher genome size, a high mutation rate (10−4) substitutions per site per year) caused by a low fidelity viral polymerase) and a high recombination frequency (up to 25 per cent for the entire genome in vivo).[8] SARS-CoV-2 may have resulted in the emergence of cross-species transmission of recombination events among a bat SARS-like Corona virus and a pangolin Corona virus.[9], [10] As the genome of SARS-CoV-2 evolved (via random mutations), mutant specimens of the virus (i.e. genetic variants) that were found to be more transmissible were naturally selected. Some SARS-CoV-2 genotypes are of concern because they maintain (or even improve) their replication fitness despite increased population immunity.[8], [10], [11], [12], [13] CoV genome mutations create virus variations ([Figure 1]), and closely similar variants are categorized into major clades/subtypes (a group of organisms that originate from a common ancestor), namely L, V, S, G, GH, GR, GV, GRY, and O ([Figure 2]) as per the Global initiative on sharing all influenza data (GISAID) nomenclature system.[8], [11], [12], [13] Such clades L, to which other SARS-CoV-2 virus strain has emerged to S, V, G, GH, GR, and GV strains ([Table 1]). However, according to Nextclade (a tool that identifies differences between text sequences and a reference sequence used by Nextstrain to assign the clades), the representation of the SARS-CoV-2 clade and its sub-variant is relatively more conclusive and clearly mention that omicron has emerged from the S clade (20B).[5] ([Figure 3]).

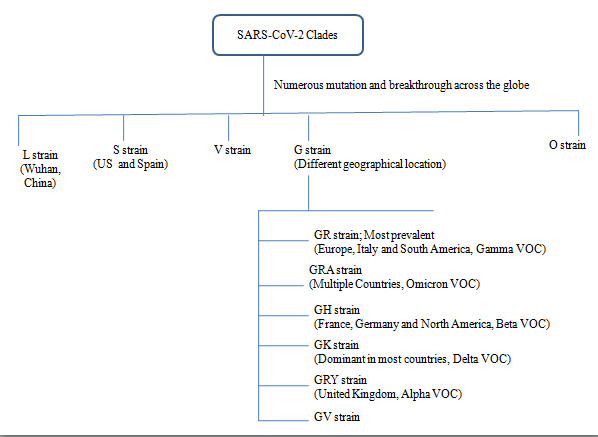
Following the rapid spread of clades G and its lineages ([Table 2]) such as B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), B.1.617 2 (Delta) and B1.1.529 (Omicron), there is a rise in the other variants, selected by growing population-level immunity and other yet undiscovered causes.[11], [13] These clades and their lineage are geographically and chronologically dispersed. For example, most of the variations that existed at the start of the epidemic are no longer present.[14] Over 78 per cent of North American isolates are from the GH and S clades, while over 65 per cent of European Union (EU) isolates are from the G and GR clades.[15]
|
Major Clades |
Reported mutation |
Global Prevalence (%) |
Remark |
Reference |
|
L |
Native Wuhan strain GenBank accession number: NC_045512, GISIAD accession ID: EPI_ISL_402124 |
1.2 |
The strain from which all it began and mutated to different clades nearly nowhere or nearly extinct. It is dominated the early stages of the pandemic |
|
|
L84S mutation in NS8 |
2.1 |
Clade S has been declining |
||
|
V |
L37F mutation in NSP6 and G251V mutation in NS3 |
1.5 |
Clade V are likely extinct |
|
|
G |
D614G mutation in the spike protein (S) |
15.8 |
Account for most common SARS-CoV-2 sequences |
|
|
GH (Beta) |
NS3-Q57H along with D614G |
21.4 |
Most common in North America (59.0%) |
|
|
GR (Gamma) |
N-G204R along with D614G |
34.0 |
Most frequently identified in four continents namely Africa (41.1%), Asia (52.7%), Oceania (74.8%), and South America (66.8%) |
|
|
GV |
S-A222V along with D614G |
22.3 |
In Europe, both GR (35.5%) and GV (34.6%) predominated |
|
|
O |
Genomes mutation which don’t cluster into any of the clades |
1.7 |
Variants of SARS-CoV-2 and the reported mutation in these variants
All viruses’ genomes accumulate mutations over time, and variants are formed. A variety of factors which include replication errors, nucleic acid damage, and editing of the genetic material by host-encoded proteins or by specialized molecular systems such as diversity-generating retro-elements (DGRs), affect the mutation rates, the viral dynamics within and among individual hosts and so it's consequences for transmission and disease in the host population progress.[16], [17] A combinations of these variables governs the onset and rapid spread of viral variants as well as the development of pandemics. The genomes of RNA viruses are highly sensitive to conversion.[18] Since SARS-CoV-2 is a positive-sense single-stranded RNA virus, its genome is expected to evolve at a rate of 1.87 x 10-6 nucleotide substitutions per site per day.[19] Within a lineage, around 20 genetic mutations per year occur across the SARS-CoV-2 genomes of 30,000 base-pair.[18] Not all mutations will survive long enough to be included in this calculation of the SARS-CoV-2 molecular clock. As the virus spreads, it gains additional opportunities for mutation and may become more difficult to eradicate. These changes can be analyzed by looking at either physical features or changes in genetic coding from one variant to another.[16] These changes are thought to be of particular concern due to their greater transmissivity rate, enhanced pathogenicity, or lower vaccine effectiveness.[20], [21] These mutations contribute to the Covid-19 pandemic’s persistence.
To determine the prehistoric human Corona virus type, Chinese researchers examined early SARS-CoV-2 viral genome sequences with bat and pangolin Corona virus strains. The identified prehistoric genome type was called "S," and it’s predominately derived from was labeled "L" to imply mutant amino acid alterations. [22] Similar investigations were conducted concurrently by western academics, however, the developed type was referred to as "B", and the ancestral type was given the designation "A". [22] The B-type mutated into other types, including B.1, which is the ancestor of the major global VOC, which were named Alpha (B.1.1.7), Beta (B.1.351 lineage), Gamma variation (P.1, P.1.1, P.1.2), Delta (B.1.617.2), and Omicron (B.1.1.529) by the WHO in 2021. [1], [2] The WHO has designated labels for notable variants based on the Greek alphabet.[2] Each variant has many names based on the nomenclature used by various evolutionary classification schemes.[5] A variant of interest (VOI) is one that possesses mutations that are suspected or known to cause significant alterations and is widely distributed (e.g., known to cause numerous clusters of diseased persons or found in many countries).[2], [5], [23] If a VOI is known to spread more quickly, cause more severe disease, escape the body’s immune response, change the clinical presentation, or reduce the effectiveness of available tools – such as public health measures, treatments, diagnostics, and vaccines, it becomes a VOC.[2], [5], [23] ([Table 2]). VOC has been raised about a strain of Covid-19 that has been more infectious and more likely to cause breakthroughs or re-infections in people who have been vaccinated or previously infected.[5] These variations are more likely to cause severe disease, remain undetected, and resist antiviral treatment. The establishment and spread of such variants are determined by two key factors: the individuals in whom the mutations occurred and what occurs later as the virus spreads between individuals. Detecting VOCs as they emerge allows for the earlier implementation of control measures. SARS-CoV-2 variant with changes in the genome that affect virus characteristics, with some indication that it may affect badly in future, but there is currently no evidence of phenotypic or epidemiological impact, necessitating increased monitoring and repeat assessment until new evidence becomes available is designated as Variants under Monitoring (VUM).[2], [5], [24]
The emergence of SARS-CoV-2 variants with high transmission rates increases the virus's ability to acquire new traits. The countries reporting variants of concern are increasing drastically.[25], [26] Viral transmissibility, disease severity, and vaccine efficacy are three major concerns with SARS-CoV-2 VOCs. When it comes to viral transmissibility, the research shows that all VOCs are more transmissible than wild-type virus. On December 18, 2020, the Alpha (B.1.1.7) variation was identified as a VOC in the United Kingdom (UK)[27] and has a significant transmission advantage over previously circulating variants, according to numerous statistical and mechanistic modeling methodologies, with estimates ranging between 40 and 80% more transmissible and than previously circulating variants.[5], [28] After being determined to have similar transmissibility to B.1.1.7, the Delta (B.1.617.2) variant, which was initially common in India, was designated a VOC in the UK on May 6, 2021.[5], [20] Beta (B.1.351) can circumvent natural immunity from earlier infection, according to findings from neutralization experiments, vaccination clinical trials, and observational analyses of population-level surveillance data.[5], [28]
In vitro study reported that the Beta and, to a fewer extent, the Gamma (P.1) variations were linked to immune evasion and spread locally but never worldwide.[29] The Alpha and Delta VOCs, on the other hand, have a global dispersion and have been linked to significant waves of illnesses and increased reproduction numbers.[30] Both variants have alterations in the polybasic furin cleavage site at position 681 (a histidine in Alpha and an arginine in Delta), linked to improved cell invasion and presumably giving an inherent transmission advantage.[30] When compared to Beta, Alpha had lesser immune evasion capabilities but higher transmission.[28] The Delta variant replaced the Alpha form, which showed increased immune evasion and furin cleavage.[30] Omicron (B1.1.529) has been identified as the fifth VOC by WHO, and it is the third (after Alpha and Delta) to achieve global domination.[5] The Omicron lineage has further split into three sub-lineages (BA.1, BA.2, and BA.3) and recombinant (XD, XE, and XF) ([Table 2]). With BA.1, it rapidly spreads over the world.[31], [32] When comparing the BA.1 Omicron genome to the native Wuhan strain, there are 30 amino acid changes in the spike glycoprotein, 15 in the receptor-binding domain (RBD) and 9 in the receptor-binding motif (RBM), the RBD sub-domain that interacts with the human angiotensin-converting enzyme-2 (ACE2) receptor.[29], [31], [32] G339D, N440K, S477N, T478K, Q498R, and N501Y are six of the mutations that boost ACE2 receptor binding affinity; Q498R and N501Y are two combinations that may improve ACE2 binding in an additive manner.[29], [30], [31], [32] In general, the Omicron RBD binds to human ACE2 with almost twice the affinity of the Wuhan RBD.[29], [31], [32] Seven Omicron RBD mutations (K417N, G446S, E484A, Q493R, G496S, Q498R, and N501Y) have been linked to decreased antibody binding, with epitopes corresponding to the three main classes of RBD-specific neutralizing antibodies dropping dramatically ([Table 2]).[5], [29], [31] The amino-terminal domain (NTD) of the Omicron spike glycoprotein contains three deletions (amino acids 69-70, 143-145, and 211) as well as an insertion (at site 214). The 69-70 deletion is likewise prevalent in the Alpha and Eta (B.1.525) variants, and it has been linked to increased fusogenicity and virions containing the cleaved spike.[30] Sub lineage of Omicron BA.1, BA.1.1, BA.2, and BA.3 has shown the major mutation in the RBD region. The recently discovered BA.2.10 and BA.2.12 sub-lineage of Omicron BA.2 in Delhi, India, has reported a 30-90% growth advantage per week over BA.2 for BA.2.12.[33] BA.2.12.1, a descendant from BA.2.12 revealed the presence the mutation namely L452Q and S704L than BA.2.[34] Other descendant of BA.2 namely BA.2.10.1, BA.2.75, BA.2.75.2, BJ.1, BA.3, BA.4, (having sub variant of BA.4.6, BA.4.7) and BA.5 (having sub lineage of BQ.1, BQ.1.1, BF.7) and sub variant of BA.5.9 has also been evolved ([Figure 7]). However, the recombinant mutant of Omicron, namely XBB, XD, XF and XE had developed when infected with two or more variants at the same time so that their genetic material is mixed by crossing over mechanism. XBB is a recombinant of BA.2.10.1 and BA.2.75 sub lineages with a global prevalence of 1.3% and detected in 35 countries.[35] XD and XF (generally called as Deltacron) are the recombinants of Delta and Omicron BA.1, where the genetic sequence is mostly the same as Delta but with aspects of the spike protein from Omicron BA.1 ([Table 2]). Another recombinant XE which was first detected in the UK on January 19, 2022, is the recombinant product of BA.1 and BA.2 and accounts for a small fraction of nearly 1100 confirmed sequences with an extremely high transmissibility rate, meaning that it would become the most dominant strain in the near future, according to WHO.[36]
Pathogenesis of omicron: Viral entry mechanism
In general, the invasion of virus requires multiple factors to get into the host cell. SARS-Cov-2 variants including Omicron fused to the host cell membrane through their spike protein with the entry receptor ACE2 in the presence of trans-membrane serine protease 2 (TMPRSS2). In this attachment mechanism, ACE2 and TMPRSS2 primarily bind with spike protein and predominantly expressed by endothelial cells across the upper respiratory tract and digestive tracts. Later those variants inject their genetic materials into the host cell through that mechanism ([Figure 8]) and afterwards, replication starts and so pathogenesis symptoms of the virus have been developed in the host.[37], [38] One important thing is that the ACE2 receptor is much rarer in lung cells than in airway cells. However, protease cofactors TMPRSS2 are more abundant in lung cells than in upper airway cells in contrast to the ACE2 receptor. That is the reason Omicron pathogenesis is mostly occurs in airways above the lungs as it requires only ACE2 receptor to invade the host cells. Since the Omicron did not use TMPRSS 2, so, it did not break the membrane rather takes wholly different route through the side to enter into the host cells. Later in air ways cells, Omicron was encapsulated into an endosome as bubble. The virus emerged from the bubble and started stealing genetic material from the host cells to begin the replicating process there. In this approach, Omicron may have acquired two benefits: first, lacking TMPRSS 2, the virus infects 7–10 times more host cells via endosomes, and second, it multiplied swiftly in the airway above the lungs, which likely assisted in its dissemination from person to person by coughing and sneezing.[37] However, other variants infect the lungs cells as it requires both ACE2 and TMPRSS2 to get encroach the genome into host cells.
A possible explanation for this can be the way the virus attacks respiratory tract cells: A few distinct factors are necessary for the virus to enter the cells. It binds to the ACE2 receptor, which is present on cells in the upper respiratory tract in particular (i.e., the throat and nasal mucosa). In lung cells, the ACE2 receptor is substantially less common.[37], [38] However, cofactors such the protease TMPRSS2 is also necessary for infection of the cells. The TMPRSS2 receptor is more prevalent in lung cells than upper airway cells, in contrast to the ACE2 receptor.
It is now conceivable to think that the virus's Omicron form has modified itself to be able to bind to the ACE2 receptor strongly through alterations in the spike protein.[37], [38] As a result, it spreads much more effectively in the upper respiratory system, making it significantly more contagious than earlier varieties. It's also possible that the virus is no longer able to employ the TMPRSS2 cofactor.[37], [38] That is the reason, the Omicron variety would have a harder time infecting lung cells, which have a limited number of ACE2 receptors and are consequently extremely reliant on the TMPRSS2 cofactor. This would therefore clarify why only a small percentage of those who contract Omicron go on to develop serious lung illness. Despite the possibility that this is good news, we regrettably cannot grant the all-clear.[37], [38] Even a considerably smaller percentage of seriously ill individuals would nevertheless place a significant load on many people and the healthcare system as a whole due to the enormous number of cases in the current "Omicron wave."
|
WHO Designation |
Lineage/ Recombinant |
First report |
Status |
Reported mutations/Feature |
Pictorial Illustration |
Reference |
|
Alpha |
B1.1.1.7 |
September 2020, UK |
VOC |
23 mutations: 13-non synonymous mutations, 4 deletions and 6 synonymous mutations. |
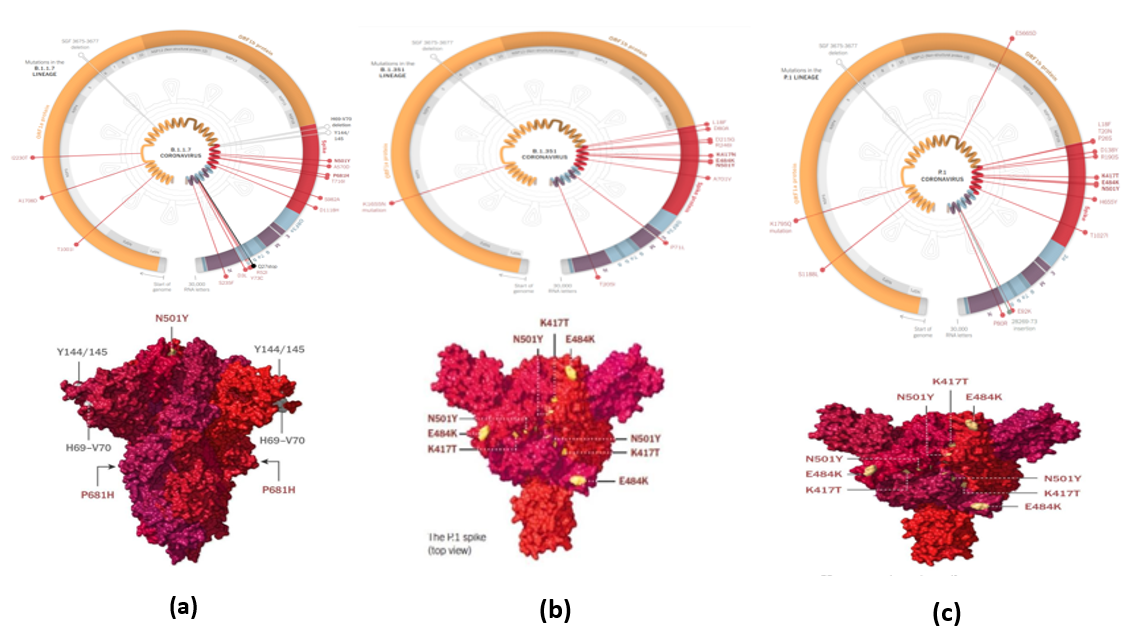
[Figure 4] (a) |
|
|
ORF1ab: |
||||||
|
Nucleotide: C3267T, C5388A, T6954C, 11288-11296 deletion. |
||||||
|
Amino acid: T1001I, A1708D, I2230T, SGF 3675-3677 deletion |
||||||
|
ORF8: |
||||||
|
Nucleotide: C27972T, G28048T, A28111G |
||||||
|
Amino acid: Q27 Stop, R52I, Y73C. |
||||||
|
Spike: |
||||||
|
Nucleotide: 21765-21770 deletion, 21991-21993 deletion, A23063T, C23271A, C23604A, C23709T, T24506G, G24914C |
||||||
|
Amino acid: |
||||||
|
HV 69-70 deletion, Y144, N501Y, A570D, P681H, T716I, S982A, D1118H deletion |
||||||
|
N: |
||||||
|
Nucleotide: 28280 GAT→CTA, C28977T |
||||||
|
Amino acid: D3L, CS235F |
||||||
|
6 synonymous mutations |
||||||
|
ORF1ab: |
||||||
|
C913T, C5986T, C14676T, C15279T, C16176T |
||||||
|
M: |
||||||
|
T26801C |
||||||
|
Beta |
B.1.351 |
May 2020, South Africa |
VOC |
ORF1ab: |
[Figure 4] (b) |
|
|
Nucleotide: C1059T, G5230T, C8660T, C8964T, A10323G, G13843T, C14408T1, C17999T. |
||||||
|
Amino acid: T265I, K1655N, H2799Y, S2900L, K3353R, D4527Y, P4715L, T5912I |
||||||
|
ORF3a: |
||||||
|
Nucleotide: G25563T, C25904T. |
||||||
|
Amino acid: Q57H, S171L |
||||||
|
Spike: |
||||||
|
Nucleotide: C21614T, A21801C, A22206G, G22299T, G22813T, G23012A, A23063T, A23403G1, G23664T |
||||||
|
Amino acid: L18F, D80A, D215G, R246I, K417N, E484K, N501Y, D614G, A701V |
||||||
|
N: |
||||||
|
Nucleotide: C28887T |
||||||
|
Amino acid: T205I |
||||||
|
E: |
||||||
|
Nucleotide: C26456T |
||||||
|
Amino acid: P71L |
||||||
|
Gamma |
P.1 |
November 2020 Brazil |
VOC |
ORF1ab: |
[Figure 4] (c) |
|
|
Amino acid: synT733C, synC2749T, S1188L, K1795Q, del 11288-11296 (3675-3677 SGF), synC12778T, synC13860T, E5665D |
||||||
|
ORF8: |
||||||
|
Amino acid: E92K,ins 28269-28273 |
||||||
|
Spike: |
||||||
|
Amino acid: L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, H655Y, T1027I |
||||||
|
N: |
||||||
|
Amino acid: P80R |
||||||
|
Delta |
B.1.617.2 |
October 2020, India |
VOC |
ORF1b: |
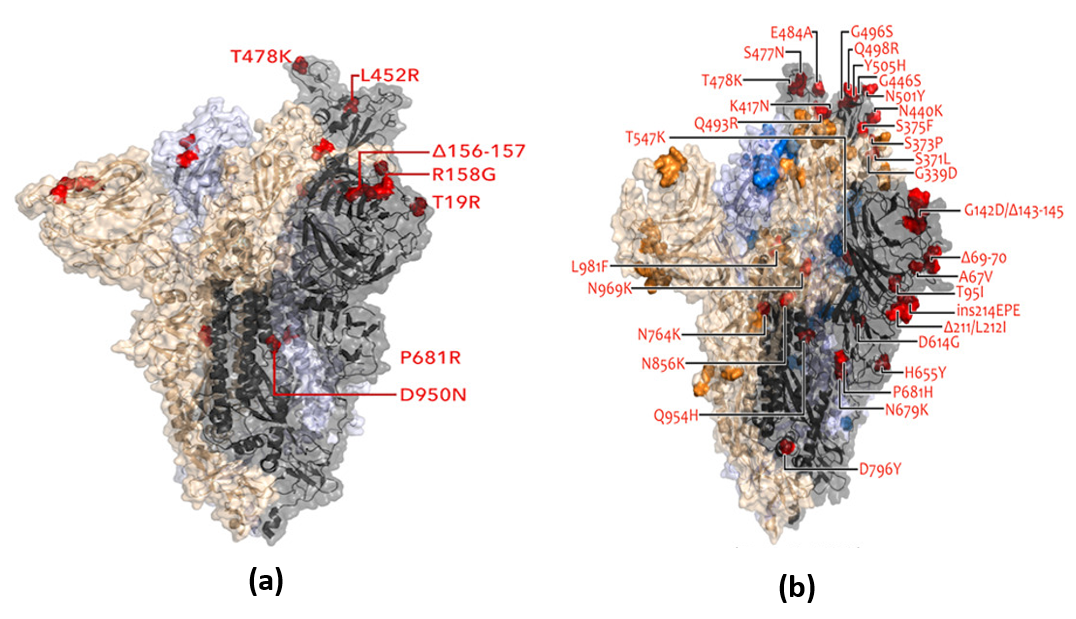
[Figure 5] (a) |
|
|
Amino acid: P314L, P1000L |
||||||
|
ORF3a: |
||||||
|
Amino acid: S26L |
||||||
|
ORF7a: |
||||||
|
Amino acid: V82A,T120I |
||||||
|
Spike: |
||||||
|
Amino acid: G142D, T19R, R158G, L452R, T478K, D614G, P681R, D950N, E156del, F157del |
||||||
|
M: |
||||||
|
Amino acid: I82T |
||||||
|
N: |
||||||
|
Amino acid: D63G, R203M, D377Y |
||||||
|
Omicron |
B1.1.529
|
November 2021, South Africa |
VOC |
ORF1ab: |
[Figure 5] (b) |
|
|
Amino acid: nsp3: K38R, nsp3: V1069I, nsp3: Δ1265, nsp3: L1266I, nsp3: A1892T, nsp4: T492I, nsp5: P132H, nsp6: Δ105-107, nsp6: A189V, nsp12: P323L,nsp14: I42V |
||||||
|
Spike: |
||||||
|
Amino acid: A67V, Δ69-70, T95I, G142D, Δ143-145, Δ211, L212I, ins214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S,S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F |
||||||
|
E: |
||||||
|
Amino acid: T9I. |
||||||
|
M |
||||||
|
Amino acid: D3G,Q19E,A63T |
||||||
|
N: |
||||||
|
P13L, Δ31-33, R203K, G204R |
||||||
|
BA.1 |
B1.1.529 |
November 2021, South Africa |
VOC Omicron Sub lineage |
Unique Mutation in Spike: |
[Figure 6] (a) |
|
|
A67V, ins214EP, R216E, S371L, G496S, T547K, N856K and L981F |
||||||
|
Spike: |
||||||
|
S375F, S37IL, S373P, G339D, N440K, N501Y, G446S, S477N, Q498R, G496S, Q493R, E484A, T478K, K417N, Y505H, K417N |
||||||
|
Common Mutations: |
||||||
|
G142D, G339D, S373P, S375F, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
||||||
|
Others: V213R, L212V, Y144del, Y145del, V143del, H69del, V70del, T95I, N211I, G446S |
||||||
|
BA.1.1 |
B1.1.529.1.1 |
January 2022, South Africa |
VOC Omicron Sub lineage |
Unique Mutation in spike: |
[Figure 6] (b) |
|
|
R346K |
||||||
|
Spike: S375F, S37IL, S373P, G339D, N440K, N501Y, G446S, S477N, Q498R, G496S, Q493R, E484A, T478K, K417N, Y505H, R346K, K417N |
||||||
|
Common Mutations: |
||||||
|
G142D, G339D, S373P, S375F, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
||||||
|
Others: |
||||||
|
V213R, L212V, Y144del, Y145del, V143del,H69del, V70del,T95I, N211I, G446S, A67V, ins 214EP, R216E, S371L, G496S, T547K, N856K, L981F, A67V, ins 214EP, R216E, S371L, G496S, T547K, N856K, L981F |
||||||
|
BA.2 |
B.1.1.529.2 |
November 2021 South Africa |
VOC Omicron Sub lineage |
Unique Mutations in spike: |
[Figure 6] (c) |
|
|
T19I, L24del deletion, P25del, P26del, A27S, V213G, T376A, R408S |
||||||
|
Spike: |
||||||
|
S371F, G339D, S373P, S375F, N440K, Q498R, N501Y,Q493R, E484A, T478K, S477N, R408S, D405N, 1376A,Y505H, K417N, E484A |
||||||
|
Same as Wild Type: |
||||||
|
G142D, G339D, S373P, S375F, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
||||||
|
Others: |
||||||
|
S371F, D405N |
||||||
|
BA.2.10 and BA.2.12 |
Sub variant of BA.2 |
April, 2022, India |
VOC Omicron Sub lineage |
BA.2.12 has growth advantage of 30-90% per week over BA.2 and come to know slowly about rest of the features and reported mutation |
||
|
BA.2.10.1 |
Sub-lineage of BA.2.10 |
2022, India |
VOC Omicron Sub lineage |
Mutations in addition to BA.2.10: |
||
|
S:V83A, H146Q, Q183E, V213E, G339H, R346T, L368I, V445P, G446S, V483A, F490V, G798D, S1003I |
||||||
|
ORF1a:K47R |
||||||
|
ORF1b:G662S |
||||||
|
M:D3Y |
||||||
|
ORF7a:I110T |
||||||
|
N:T282I |
||||||
|
nucleotide:C15738T, T15939C, T17859C, G28079T |
||||||
|
S: V445P is a 2-nucleotide mutation (GTT to CCT) |
||||||
|
BA.2.12.1 |
Descendant of the BA.2.12 |
2021, United States |
VOC Omicron Sub lineage |
Two mutation in L452Q and S704L than BA.2 |
||
|
BA.2.75 |
Sub variant of BA.2 |
2022, India |
VOC Omicron Sub lineage |
Presence of two distinct mutations; The first is G446S, a location that contributes significantly to this variant's resistance to antibodies and the second mutation is R493Q, which permits the virus to connect to ACE2 receptors, allowing it to adhere to cells more easily |
||
|
BJ.1 |
Descendant of Omicron BA.2 |
2022, Austria |
VOC Omicron Sub lineage |
Mutations in the spike protein S:V83A, H146Q, Q183E, V213E, G339H, R346T, L368I, V445P, G446S, V483A, F490V, G798D, S1003I |
A second generation variant |
|
|
BA.3 |
B.1.1.529.3 |
March, 2022 North West South Africa |
VOC Omicron Sub lineage |
Unique Mutation in spike: |
[Figure 6] (d) |
|
|
R214del |
||||||
|
Spike: |
||||||
|
S371F, G339D, S373P, S375F, N440k, G446S, Q493R, K417N, E484A, T478K, S477N, N501Y, S371F D405N |
||||||
|
Common Mutations: |
||||||
|
G142D, G339D, S373P, S375F, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
||||||
|
Others: |
||||||
|
Y144del, H69del, V70del, T951, Y145del, N2111, L212V, V213R, R214del, V143del |
||||||
|
BA.4 |
More similar to BA.2 |
January and February 2022, South Africa |
VOC Omicron Sub lineage |
Additionally L452R, F486V, and R493Q compared to BA.2 |
||
|
BA.4.6 |
Descendant of the BA.4 |
January and February 2022, South Africa
|
VOC Omicron Sub lineage |
Contain the additional mutations R346T and N658S |
||
|
BA.4.7 |
Descendant of the BA.4 |
January and February 2022, South Africa |
VOC Omicron Sub lineage |
Additional mutation R346S |
||
|
BA.5 |
More similar to BA.2 |
April 2022, South Africa |
VOC Omicron Sub lineage |
Additionally L452R, F486V, and R493Q compared to BA.2 |
||
|
BA.5.9 |
Descendant of the BA.5 |
April 2022, South Africa |
VOC Omicron Sub lineage |
Additional mutation R346I |
||
|
BQ.1 |
Sub lineage of BA.5 |
December 2022, United State |
VOC Omicron Sub lineage |
Carries spike mutations in some key antigenic sites, including K444T and N460K |
||
|
BQ.1.1 |
Sub lineage of BA.5 |
December 2022, United State |
VOC Omicron Sub lineage |
Five convergent substitutions R346T, K444T, L452R, N460K, and F486V |
||
|
BF.7 |
BA.5.2.1.7 |
October 2022, China |
VOC Omicron Sub lineage |
Unique mutation R346T |
||
|
XD and XF |
Recombinants of Delta and Omicron BA.1 |
December 2021, France Denmark and Belgium |
Recombinant Variant |
XD has the BA.1 Spike protein as well as the rest of the genome from Delta structural protein |
|
|
|
XBB |
BA.2.10.1 and BA.2.75 sub lineages |
Detected in 35 countries |
Recombinant Variant |
|||
|
XE |
Recombinants of BA.1 and Omicron BA.2 |
January 2022, UK |
Recombinant Variant
|
It has three mutations that are not present in all BA.1 or BA.2 sequences. XE had reported a 9.8 percent higher growth rate than BA.2 and shown community transmission. |
||
|
Lamda |
C.37 |
December 2020, Peru |
VOI |
ORF1ab: |
||
|
PLpro: T428I, PLpro: P1469S, PLpro: F1569V, nsp4:L438P, nsp4:T492I, 3CL: G15S, nsp6:S106K, nsp6:Δ107-109, RdRP: P323L |
||||||
|
Spike: |
||||||
|
Amino acid: G75V, T76I, L452Q, F490S, D614G, R246N, T859N, Δ246-252 |
||||||
|
N: |
||||||
|
P13LN, R203KN, G204RN, G214C |
||||||
|
Mu |
B.1.621 |
January 2021, Colombia |
VOI |
ORF1a: |
||
|
Amino acid: T1055A, T1538I, T3255I, Q3729R |
||||||
|
ORF1b: |
||||||
|
Amino acid: P314L, P1342S |
||||||
|
ORF3a: |
||||||
|
Amino acid: Q57H, del 257/257 |
||||||
|
ORF8: |
||||||
|
Amino acid- T11K, P38S, S67F |
||||||
|
Spike: |
||||||
|
Amino acid: T95I, Y144S, Y145N, R346K, E484K, N501Y, D614G, P681H, D950N |
||||||
|
N: |
||||||
|
Amino acid: T205I |
||||||
|
Epsilon |
B.1.427 |
March 2020, USA |
VOI |
ORF1ab: |
||
|
nsp2:T85I, nsp4:S395T, RdRP: P323L, nsp13:P53L, nsp13:D260Y |
||||||
|
ORF3a: |
||||||
|
Q57H |
||||||
|
Spike: |
||||||
|
L452R |
||||||
|
N: |
||||||
|
T205I. |
||||||
|
Others: |
||||||
|
S13I, W152C, D614G |
||||||
|
Zeta |
P.2 |
April 2020, Brazil |
VOI |
ORF1ab: |
||
|
3CL: L205V, nsp7:L71F, RdRP: P323L |
||||||
|
Spike: |
||||||
|
E484K |
||||||
|
N: |
||||||
|
A119S, R203K, G204R, M234I |
||||||
|
Others: |
||||||
|
D614G, V1176F |
||||||
|
Eta |
B.1.525 |
December 2020, UK and Nigeria, |
VOI |
ORF1ab: PLpro: T1189I, nsp6: S106K, nsp6:Δ107-109, RdRP: P323 ORF6: F2H, Δ3 In spike RBD: E484K E: L21F M: I82T N: S2Y, Δ3, A12G, T205I. Others: Q52R, A67V, Δ69-70, Δ144-145, ΔH69/ΔV70 deletion, D614G, Q677H, F888L |
||
|
Theta |
P.3 |
January 2021, Philippines |
VOI |
ORF1ab: |
||
|
Amino acid: F924F, D1554G, S2433S, L3201P, D3681E, N3928N, L3930F, P4715L, A5692V |
||||||
|
ORF1a: |
||||||
|
Amino acid: D1554G, S2625F, D2980N, L3201P, D3681E, L3930F |
||||||
|
ORF1b: |
||||||
|
Amino acid: P314L, L1203F, A1291V |
||||||
|
ORF8: |
||||||
|
Amino acid: K2Q |
||||||
|
Spike: |
||||||
|
Amino acid: LGV141, 143del, E484K, N501Y, G593G, D614G, P681H, S875S, E1092K, H1101Y, V1176F |
||||||
|
N: |
||||||
|
Amino acid: R203K, G204R |
||||||
|
Iota |
B.1.526 |
November 2020, United State |
VOI |
ORFLab: |
||
|
nsp6:Δ106-108 |
||||||
|
Spike: |
||||||
|
E484K |
||||||
|
Others: |
||||||
|
L5F, T95I, D253G, D614G and A701V, S477N |
||||||
|
Kappa |
B.1.617. |
December 2020, India |
VOI |
ORF1ab: |
||
|
Nucleotide: C3457T, C4957T, A11201G, G17523T, A20396G. |
||||||
|
Amino acid: T1567I, T3646A, M5753I, K6711R, P314L, G1129C, M1352I, K2310R, S2312A |
||||||
|
ORF3a: |
||||||
|
Nucleotide: C25469T. |
||||||
|
Amino acid: S26L |
||||||
|
ORF1a: |
||||||
|
Amino acid: T1567I, T3646A |
||||||
|
ORF1b: |
||||||
|
Nucleotide: T27638C |
||||||
|
Amino acid: V82A |
||||||
|
Spike: |
||||||
|
Nucleotide: T21895C, T21895C, T22917G, G23012C, C23604G |
||||||
|
Amino acid: E154K, L452R, E484Q, D614G, P681R, Q1071H |
||||||
|
N: |
||||||
|
Nucleotide: G28881T |
||||||
|
Amino acid: R203M, D377Y |
||||||
|
M: |
||||||
|
Amino acid: I82S |


Conclusion
Corona virus infection is a growing global health hazard that has infected a large section of the global population. The explanation for the appearance of VOCs and the biological and evolutionary aspects of the novel variant of concern omicron is examined in this paper. Based on the evidence of various modifications that could impact its actions, the WHO has identified Omicron as a variant of concern. There is consistent evidence that Omicron spreads substantially faster than Delta in regions where community transmission is established, with a doubling duration of 2-3 days. Also, a number of sub-lineages of omicron are also spreading enormously. The overall danger of this new variant remains extremely high. Since Omicron's debut, scientists have been on high alert and trying to learn more about the characteristics of this new variant; because many of these changes have never been seen in any virus strain previously. Even if the Covid-19 vaccines do not give 100% protection against this new variant, each of them can decrease severe illnesses and death rates up to a certain extent. As a result, encouraging vaccines, boosters, and accessible testing will play a critical role in preventing disease spread and quickly detecting outbreaks. However, people who have received their Covid-19 immunizations and are exposed to Covid-19 are less likely to suffer severe illness than those who have not received their Covid-19 vaccines and are exposed to Covid-19. Another thing to consider is that Omicron is more likely to re-infect people due to its rapid transmission rate. After the third dose, the effect of vaccinations against Covid-19 has yet to be determined. At this time, there is no additional proof that existing immunizations do not work on Omicron; nevertheless, some of the newly found Spike gene abnormalities may impair the efficiency of existing vaccines. We have preliminary information that implies Omicron is less severe than Delta. However, if there are more cases, there will be more hospitalizations, and if the healthcare system is overburdened, individuals will die because they will not receive the proper care. Emerging omicron subtypes are also a source of worry. Nowadays, according to research, the Omicron coronavirus variant may minimize the prevalence of severe Covid-19 disease, making the infection less disruptive to individuals and society in the future. There is, however, no proof that this is the case. Few reports are circulating these days about introducing omicron-specific vaccines as a long-term help against Covid-19; the assumption is that an omicron-specific booster would have high vaccine effectiveness against infection, but the choices are still being researched. It's also vital to remember that all SARS-CoV-2 variants, including the Delta form, can cause serious illness or death, which is why avoiding the virus's transmission and minimizing your chance of infection are so critical.
Conflict of Interest
None.
Source of Funding
None.
References
- C Jonathan, Z Carl. Tracking Omicron and Other Corona virus Variants. 2022. [Google Scholar]
- C Jonathan, Z Cart. Inside the B.1.1.7 Coronavirus variant. 2021. [Google Scholar]
- P Roger. SARS-COV-2 Variants: Clinical and Epidemiological Implications. . [Google Scholar]
- . UNICEF. What we know about the Omicron variant, What is Omicron and what precautions should you take to protect your family? . Updated on 14 January 2021. [Google Scholar]
- . World Health Organization (WHO). Classification of Omicron (B.1.1.529): SARS-CoV-2 variant of concern. . [Google Scholar]
- . World Health Organization (WHO). WHO advice for international traffic in relation to the SARS-CoV-2 Omicron variant (B.1.1.529). . [Google Scholar]
- E Cameroni, JE Bowen, LE Rosen, C Saliba, SK Zepeda, K Culap. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 2022. [Google Scholar]
- . World Health Organization (WHO) Tracking SARS-CoV-2 variants. . [Google Scholar]
- M Mohiuddin, K Kasahara. Investigating the aggressiveness of the COVID-19 Omicron variant and suggestions for possible treatment options. Respir Med 2021. [Google Scholar] [Crossref]
- . India Today, 2021, December 26, The new COVID variant Omicron is now present in nearly 115 countries. . [Google Scholar]
- J Singh, P Pandit, AG Mcarthur, A Banerjee, K Mossman. Evolutionary trajectory of SARS-CoV-2 and emerging variants. Virol J 2021. [Google Scholar]
- PH Tsai, ML Wang, DM Yang, KH Liang, SJ Chou, SH Chiou. Genomic variance of Open Reading Frames (ORFs) and Spike protein in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). J Chin Med Assoc 2020. [Google Scholar]
- H Guo, BJ Hu, XL Yang, LP Zeng, B Li, S Ouyang. Evolutionary arms race between virus and host drives genetic diversity in bat severe acute respiratory syndrome-related corona virus spike genes. J Virol 2020. [Google Scholar]
- A Sengupta, SS Hassan, PP Choudhury. Clade GR and clade GH isolates of SARS-CoV-2 in Asia show highest amount of SNPs. Infect Genet Evol 2021. [Google Scholar] [Crossref]
- C Roemer, EB Hodcroft, RA Neher, T Bedford. SARS-CoV-2 clade naming strategy for 2022. . [Google Scholar]
- SM Hamed, WF Elkhatib, AS Khairalla, AM Noreddin. Global dynamics of SARS-CoV-2 clades and their relation to COVID-19 epidemiology. Sci Rep 2021. [Google Scholar]
- . What’s the difference between mutations, variants and strains? A guide to Covid terminology. 2021. [Google Scholar]
- I Aksamentov, C Roemer, EB Hodcroft, RA Neher. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Sour Software 2021. [Google Scholar]
- MK Parvez, S Parveen. Evolution and emergence of pathogenic viruses: past, present, and future. Intervirology 2017. [Google Scholar]
- R Sanjuán, P Domingo-Calap. Mechanisms of viral mutation. Cell Mol Life Sci 2016. [Google Scholar]
- SP Otto, T Day, J Arino, C Colijn, J Dushoff, M Li. The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic. Curr Biol 2021. [Google Scholar]
- S Wang, X Xu, C Wei, S Li, J Zhao, Y Zheng. Molecular evolutionary characteristics of SARS-CoV-2 emerging in the United States. J Med Virol 2022. [Google Scholar]
- . SARS-CoV-2 variants of concern and variants under investigation in England - Technical briefing 29. (PDF). UK Health Security Agency. 2021. [Google Scholar]
- N Shahhosseini, G Babuadze, G Wong, GP Kobinger. Mutation signatures and in silico docking of novel SARS-CoV-2 variants of concern. Microorganisms 2021. [Google Scholar]
- BBO Munnink, N Worp, DF Nieuwenhuijse, RS Sikkema, B Haagmans, RA Fouchier. The next phase of SARS-CoV-2 surveillance: real-time molecular epidemiology. Nat Med 2021. [Google Scholar]
- . SARS-CoV-2 Variant Classifications and Definitions. [internet]. Updated on 5 June 2021. [Google Scholar]
- . World Health Organization. Corona virus disease (COVID-19) pandemic. 2021. [Google Scholar]
- L Dyson, EM Hill, S Moore, J Curran-Sebastian, MJ Tildesley, KA Lythgoe. Possible future waves of SARS-CoV-2 infection generated by variants of concern with a range of characteristics. Nat Commun 2021. [Google Scholar]
- . Public Health England (PHE): Investigation of novel SARS-CoV-2 variant. Variant of concern 20201/01.2020. . [Google Scholar]
- NG Davies, S Abbott, RC Barnard, CI Jarvis, AJ Kucharski, JD Munday. Estimated transmissibility and impact of SARS-CoV-2 lineage B. 1.1. 7 in England. Science 2021. [Google Scholar] [Crossref]
- L Lin, Y Liu, X Tang, D He. The disease severity and clinical outcomes of the SARS-CoV-2 variants of concern. Front Public Health 2021. [Google Scholar]
- R Viana, S Moyo, DG Amoako, H Tegally, C Scheepers, CL Althaus. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 2022. [Google Scholar]
- BJ Willett, J Grove, OA Maclean, C Wilkie, G DeLorenzo, W Furnon. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat Microbiol 2022. [Google Scholar]
- K Spencer. The latest Covid variant is 1.5 times more contagious than omicron and already circulating in almost half of U.S. state. Health and Science. CNBC 2022. [Google Scholar]
- . Omicron BA.2: What we know about the Covid sub-variant. 2022. [Google Scholar]
- . Eight New Variants Detected in Delhi Samples, Omicron Sub-Lineage BA.2.12 Dominant. 2022. [Google Scholar]
- P Qu, J Faraone, JP Evans, X Zou, YM Zheng, C Carlin. Neutralization of the SARS-CoV-2 omicron BA. 4/5 and BA. 2.12.1 sub variants. N Engl J Med 2022. [Google Scholar]
- . World Health Organization (WHO). TAG-VE statement on Omicron sublineages BQ.1 and XBB. . [Google Scholar]
- . What Is the Omicron XE Variant and Should We Be Worried?. 2022. [Google Scholar]
- A Gupta, MV Madhavan, K Sehgal, N Nair, S Mahajan, TS Sehrawat. Extrapulmonary manifestations of Covid-19. Nat Med 2020. [Google Scholar]
- M Scudellari. Omicron’s Surprising Anatomy Explains Why It Is Wildly, Contagious. Scientific American. 2022. [Google Scholar]
- S Kumar, K Karuppanan, G Subramaniam. Omicron (BA.1) and sub-variants (BA.1.1, BA.2, and BA.3) of SARS-CoV-2 spike infectivity and pathogenicity: A comparative sequence and structural-based computational assessment. J Med Virol 2022. [Google Scholar]
- S Thakur, S Sasi, SG Pillai, A Nag, D Shukla, R Singhal. SARS-CoV-2 Mutations and Their Impact on Diagnostics, Therapeutics and Vaccines. Front Med (Lausanne) 2022. [Google Scholar] [Crossref]
- S Kumar, TS Thambiraja, K Karuppanan, G Subramaniam. Omicron and Delta variant of SARS-CoV-2: a comparative computational study of spike protein. J Med Virol 2022. [Google Scholar]
- . Variant: 21K (Omicron). 2021. [Google Scholar]
- . SARS-CoV-2 variants of concern and variants under investigation in England - Technical briefing 29. (PDF). UK Health Security Agency. 2021. [Google Scholar]
- . Spike Variants: Spike Variants: Delta variant, aka B.1.617.2. 2021. [Google Scholar]
- . Spike Variants: Spike Variants: Gamma variant, aka P.1, B.1.1.28.1, 20J/501Y.V3 variant, and K417T/E484K/N501Y. 2021. [Google Scholar]
- . Spike Variants: Kappa variant, aka B.1.617.1., covdb.stanford.edu. 2021. [Google Scholar]
- . Spike Variants: Spike Variants: Beta variant, aka B.1.351, 501Y.V2, 20C/501Y.V2, and South African COVID-19 variant 24 June 2021, covdb.stanford.edu. . [Google Scholar]
- . Spike Variants: Iota variant, aka B.1.526., covdb.stanford.edu. 2021. [Google Scholar]
- . Spike Variants: Theta variant, aka B.1.128., covdb.stanford. 2021. [Google Scholar]
- . Spike. covdb.stanford.edu. 2021. [Google Scholar]
- . Spike Variants: Zeta variant, aka P.2., covdb.stanford.edu. 2021. [Google Scholar]
- . Spike Variants: Epsilon variant, aka B.1.427/B.1.429., covdb.stanford.edu. 2021. [Google Scholar]
- . Stanford Corona virus Antiviral & Resistance Database. SARS-CoV-2 variants. 2021. [Google Scholar]
- . Spike Variants: Lambda variant, aka B.1.1.1., covdb.stanford.edu. 2021. [Google Scholar]
- . UK: XE may have a growth rate ~10% greater than Omicron BA.2 *1. 2022. [Google Scholar]
- . World Health Organization (WHO). TAG-VE statement on Omicron sublineages BQ.1 and XBB. . [Google Scholar]
- M Sharma. Explained: What's BF.7 Omicron Sub-Variant That's Driving China's Covid Wave, What Is Its Severity?. 2022. [Google Scholar]
- S Lomte, Tarun. Characterization of SARS-CoV-2 Omicron BQ.1.1. 2022. [Google Scholar]
- . Omicron BQ.1, BQ.1.1 Cases Rising: Key Points On Symptoms, Risks And Prevention. 2022. [Google Scholar]
- Q Wang, Z Li, J Ho, Y Guo, A Y Yeh, H Mohri. Resistance of SARS-CoV-2 omicron subvariant BA. 4.6 to antibody neutralisation. Lancet Infect Dis 2022. [Google Scholar]
- RK Mohapatra, V Kandi, AK Sarangi, S Verma, HS Tuli, S Chakraborty. The recently emerged BA. 4 and BA. 5 lineages of Omicron and their global health concerns amid the ongoing wave of COVID-19 pandemic-Correspondence. Int J Surg 2022. [Google Scholar] [Crossref]
- . New Omicron variant BJ.1 – dangerous mutation. 2022. [Google Scholar]
- NS Shaheen, A Mohamed, Y Soliman, OA Abdelwahab, RA Diab, MT Desouki. Could the new BA. 2.75 sub-variant lead to another COVID-19 wave in the world?–correspondence. Int J Surg 2022. [Google Scholar] [Crossref]
- . BA.2.10.1 sub lineage with S:V83A, 144del, H146Q, Q183E, V213E, G339H, R346T, L368I, V445P, G446S, V483A, F490V, S1003I (11 seq, West Bengal and 1 in New York) #915 . . [Google Scholar]
- . Spike Variants: Alpha variant, aka B.1.1.7, 501Y.V1, 20I/501Y.V1 covdb.stanford.edu. 2021. [Google Scholar]
- . Eight New Variants Detected in Delhi Samples, Omicron Sub-lineage BA.2.12 Dominant: Sources. 2022. [Google Scholar]
